-
PDF
- Split View
-
Views
-
Cite
Cite
Mauricio González-Agüero, Leonardo Pavez, Freddy Ibáñez, Igor Pacheco, Reinaldo Campos-Vargas, Lee A. Meisel, Ariel Orellana, Julio Retamales, Herman Silva, Mauricio González, Verónica Cambiazo, Identification of woolliness response genes in peach fruit after post-harvest treatments, Journal of Experimental Botany, Volume 59, Issue 8, May 2008, Pages 1973–1986, https://doi.org/10.1093/jxb/ern069
Close - Share Icon Share
Abstract
Woolliness is a physiological disorder of peaches and nectarines that becomes apparent when fruit are ripened after prolonged periods of cold storage. This disorder is of commercial importance since shipping of peaches to distant markets and storage before selling require low temperature. However, knowledge about the molecular basis of peach woolliness is still incomplete. To address this issue, a nylon macroarray containing 847 non-redundant expressed sequence tags (ESTs) from a ripe peach fruit cDNA library was developed and used. Gene expression changes of peach fruit (Prunus persica cv. O'Henry) ripened for 7 d at 21 °C (juicy fruit) were compared with those of fruit stored for 15 d at 4 °C and then ripened for 7 d at 21 °C (woolly fruit). A total of 106 genes were found to be differentially expressed between juicy and woolly fruit. Data analysis indicated that the activity of most of these genes (>90%) was repressed in the woolly fruit. In cold-stored peaches (cv. O'Henry), the expression level of selected genes (cobra, endopolygalacturonase, cinnamoyl-CoA-reductase, and rab11) was lower than in the juicy fruit, and it remained low in woolly peaches after ripening, a pattern that was conserved in woolly fruit from two other commercial cultivars (cv. Flamekist and cv. Elegant Lady). In addition, the results of this study indicate that molecular changes during fruit woolliness involve changes in the expression of genes associated with cell wall metabolism and endomembrane trafficking. Overall, the results reported here provide an initial characterization of the transcriptome activity of peach fruit under different post-harvest treatments.
Introduction
Extended storage of peaches, nectarines and other stone fruit can negatively affect fruit quality due to the development of physiological disorders, known as chilling injury (CI) or internal breakdown (Lill et al., 1989; Lurie and Crisosto, 2005). One of the most common disorders is woolliness, which becomes apparent when fruit are ripened after storage at 2–8 °C for a period of at least 2 weeks (Ben Arie and Lavee, 1971; Lill et al., 1989). Under these conditions, fruit fail to soften normally and develop a dry, woolly texture instead of becoming juicy. This disorder is of commercial importance since shipping of peaches to distant markets and storage before selling require low temperature (Campos-Vargas et al., 2006). Additionally, it is not possible to perceive woolliness on the fruit surface. However, when the fruit is bitten into, the lack of juice results in an absence of flavour and dryness, which make it unpalatable and leads to consumer rejection (Zhou et al., 2000b).
A recurring hypothesis to explain peach woolliness supports the idea that changes in cell wall enzyme activity during cold storage will affect the metabolism of cell wall polysaccharides during the subsequent ripening at warm temperatures (Ben Arie and Lavee, 1971; Buescher and Furmanski, 1978). A characteristic of the ripening of melting-flesh peaches is an increase in the activity of cell wall-degrading enzymes that are responsible for fruit softening. As a peach ripens, several cell wall modifications, such as the solubilization or depolymerization of pectin and matrix glycans, are observed, and these changes are of considerable importance in fruit texture (Ben-Arie et al., 1979; Labavitch, 1981). Woolliness has been attributed to an imbalance between the activity of the cell wall-degrading enzymes, polygalacturonase (PG) and pectin methylesterase (PME) (Ben-Arie and Sonego, 1980; Zhou et al., 2000a, b, c; Brummell et al., 2004). Relatively high PME and low PG activity in chilling-injured fruit leads to an accumulation of de-methylesterified pectins which are not subsequently depolymerized. Ultrastructural analyses have revealed that in woolly peach the reduced pectin depolymerization results in enlarged intercellular spaces, which affect cell–cell adhesion and lead to an abnormal breakage of cells and the release of juice (Brummell et al., 2004). Furthermore, it has been proposed that the high molecular weight pectins along with cell wall calcium form a gel binding free water and contributing to the woolliness phenotype (Dawson et al., 1993; Zhou et al., 2000a). Besides, the activities of endo-1,4-β-glucanase, endo-1,4-β-mannanase, β-galactosidase, α-arabinosidase, and expansin have also been reported to decrease in woolly fruit compared with juicy fruit, leading to further alteration of cell wall metabolism (Zhou et al., 2000b; Obenland et al., 2003; Brummell et al., 2004).
Since modifications in cell wall structure during the ripening process are proposed to be key determinants of the diminished juice content in woolly fruit, the large complexity and functional diversity of cell components associated with this process should be considered. Recent transcriptome analyses of peach ripening have contributed to understandimg the role of transcriptional gene regulation during fruit softening (Trainotti et al., 2003, 2006). These studies have revealed an ordered and sequential expression of several genes encoding novel cell wall metabolism-related components, many of which were up- or down-regulated by ethylene. Even though the softening of the cell wall has been traditionally attributed to the co-operative activity of different degradation enzymes, the transcriptome analysis carried out by Trainotti et al. (2003) revealed that peach softening was accompanied by an increased expression of genes encoding structural cell wall proteins, which parallels the expression patterns of genes that code for cell wall-degrading enzymes. Thus, these analyses have uncovered new elements and cellular pathways acting during peach softening, supporting the view that remodelling of the cell wall involves a higher level of complexity than previously thought.
Under the assumption that peach woolliness is the result of an abnormal fruit ripening, a transcriptome analysis of gene expression changes associated with peach woolliness can be useful in defining the cellular processes that affect peach quality during post-harvest and can help to determine how woolliness is developed by the fruit. To address these issues, a cDNA macroarray containing 847 peach unigenes spotted onto nylon membranes was designed and used. Peaches (Prunus persica cv. O'Henry) were harvested and exposed to different treatments that simulate post-harvest conditions. The transcriptome patterns of fruit ripened for 7 d at 21 °C (juicy fruit) were compared with those stored for 15 d at 4 °C and then ripened for 7 d at 21 °C (woolly fruit). The objective of the study was to identify candidate genes that would provide new insights into the complex issue of peach fruit woolliness.
Materials and methods
Fruit samples and post-harvest conditions
Peaches (P. persica cv. O'Henry, cv. Elegant Lady, cv. Flamekist) were commercially harvested and exposed to three different treatments that simulate post-harvest conditions: (c1) 7 d at 21 °C; (c2) 15 d at 4 °C; and (c3) 7 d at 21 °C after 15 d at 4 °C. Conditions c1 and c3 were applied to obtain juicy and woolly fruit samples for macroarray experiments, while samples obtained with condition c2 allowed evaluation of the effects of cold storage on the expression of selected genes. At the end of the different treatments, 10 fruit from each condition were halved, and one-quarter was immediately frozen in liquid nitrogen and stored at –80 °C. In the case of fruit exposed to conditions c1 and c3, one-quarter was used to determine woolliness as described by Campos-Vargas et al. (2006). Fruit were classified as woolly or juicy when they contained <10% (w/w) of juice as described by Campos-Vargas et al. (2006). The three fruit from treatments c1 and c3 that presented the highest and the lowest percentage of juice, respectively, were selected for RNA extraction, macroarray hybridization, and quantitative real-time PCR experiments.
Preparation of DNA macroarrays
The source of the spotted cDNA was a ripe peach fruit (P. persica L. Batsch, cv. Loring) cDNA library. The library was constructed in the pBluescript II SK(–) vector and it was obtained from Clemson University Genomics Institute (www.genome.clemson.edu/projects/peach/est/, Jung et al., 2004). A total of 847 cDNAs were selected from this library based on the functional properties assigned to their encoded proteins using an in-house gene-ranking method. This analysis involved a sequence homology search using tBLASTx of 3843 non-redundant sequenced clones from the cDNA library against the Arabidopsis CDS database (http://www.arabidopsis.org) to generate a list of orthologues (e-value <1E-10, identity >70% over a minimum of 50 amino acids) which were further analysed by manual searches in the annotated database, gene ontology terms, and information obtained in the literature. Sequence management was performed with database tools developed in the authors' laboratory. Domain-based analyses were performed using SMART (http://smart.embl-heidelberg.de, Schultz et al., 1998) and InterPro Scan (http://www.ebi.ac.uk/InterProScan/, Quevillon et al., 2005). Signal peptide prediction and subcellular localization analyses were performed using SignalP (http://www.cbs.dtu.dk/services/SignalP, Bendtsen et al., 2004) and TargetP v1.01 software (www.cbs.dtu.dk/services/TargetP/, Emanuelsson et al., 2000).
Selected EST clones were picked from –80 °C stocks and grown overnight in 96-well plates in 200 μl of LB plus 50 μg ml−1 of ampicillin. Overnight cultures were used directly as templates in PCRs containing 10× buffer, 0.25 mM each of dATP, dCTP, dGTP, and dTTP, 2.5 U of Taq DNA polymerase, and 0.2 μM of T3 and T7 forward/reverse primers. Aliquots (5 μl) of each reaction were loaded onto a 1% (w/v) agarose gel to determine product quality. A 20 μl aliquot of the PCR products was arrayed in 96-well plates and mixed with an equal volume of 50% dimethylsulphoxide (DMSO). PCR products (70 ng) were single spotted onto nylon membranes (10×7 cm) Immobilon NY+ (Millipore, Billerica, MA, USA) using an 8-pin print-head (Arraylt model SSP015) and the arraying robot Versarray Chip Writer Compact (Bio-Rad, Hercules, CA, USA). The membranes were treated as described in González-Agüero et al. (2005), and the cDNA was fixed to the membrane by UV cross-linking using an Ultraviolet Crosslinker CL-1000 (UVP). In addition to the selected ESTs, the following controls were spotted onto membranes: (i) a fragment of the vector pBluescript II obtained by amplification with the T3 and T7 universal primers; (ii) several aliquots of 50% DMSO; (iii) Pp-Expansin (GenBank: AB029083), a gene that shows a decreased expression in woolly peach compared with a juicy peach (Obenland et al., 2003); and (iv) cDNA from a Bacillus subtilis gene dap (obtained from the ATCC; number 87486), which hybridizes to an in vitro synthesized poly(A) RNA that was added to the fruit mRNA (dilution 1/200) prior to the labelling process and that was used as spike mRNA to normalize expression data between membranes (Kane et al., 2000).
RNA extraction and mRNA isolation
Total RNA was isolated from fruit as described by Meisel et al. (2005). The quantity and quality of the RNA were assessed by measuring the A260/280 and A260/240 ratios and by electrophoresis on a 1.2% formaldehyde–agarose gel. Typical yield was 20–40 μg of total RNA mg−1 tissue. The mRNAs were purified using an Oligotex Mini kit (Qiagen, German Town, MD, USA), following the manufacturer's recommendations.
Probe preparation and macroarray hybridization
The 32P-labelled target DNA samples were prepared from poly(A)+ RNAs by incorporation of [α-32P]dCTP during first-strand cDNA synthesis, according to the protocol described by Bernard et al. (1996); unincorporated radioactive nucleotides were removed using the QIAquick Nucleotide Removal according to the manufacturer's instructions (Qiagen). The labelled cDNA products were denatured and immediately used for membrane hybridization. Hybridization conditions were as follows: pre-hybridization in 5 ml of hybridization buffer (5× SSC, 5× Denhardt solution, 1% SDS, and 50% formamide) for 1–3 h at 42 °C. Hybridization was performed in the same buffer for 16–18 h at 42 °C. Membranes were washed as described by González-Agüero et al. (2005). After washing, the membranes were sealed with a plastic film and exposed for 72–96 h to Phosphor screens (Kodak, Rochester, NY, USA).
Macroarray experimental design and data analyses
Macroarray experiments were performed in six independent labelling/hybridization events (experimental replicates) of three juicy fruit and three woolly fruit (biological replicates). Radioactive images of the 36 hybridized membranes were obtained using a scanner Personal Molecular Image FX (Bio-Rad), and quantification of the signal intensity was performed using the VersArray Analyzer software (Bio-Rad). Raw values were measured as the volume of pixels within a circle encompassing the spot. Local background values were measured in the corners of each spot and were subtracted from the signal intensity values for each spot. Spots that showed: (i) signal mean <[background mean+(1×background SD)]; (ii) qcom <0.8; and (iii) intermembrane coefficients of variation (CVs) >0.5 were considered as low-quality spots and were removed. Qcom values were calculated as described in Wang et al. (2001). Then, the net intensity value of each spot was normalized using dap, a spike mRNA control. Differentially expressed genes were defined with SAM (significance analysis of microarray) (Tusher et al., 2001) using a false discovery rate (FDR) <10%. Expressed genes were considered as those that were detected in at least two fruit, and SAM analysis was applied only if a minimum of three experimental replicates were available. For signals that fell within the background noise in one condition but in the other condition were high enough to pass the filters in the three biological replicates, a protocol described by Wu et al. (2004) was applied.
Differentially expressed genes were further analysed by determining their open reading frame (ORF) using the graphical analysis tool ORF Finder (http://www.ncbi.nih.gov/gorf/gorf.html) and analysed for similarity against the NCBI non-redundant database using the BLASTp algorithm (http://www.ncbi.nlm.nih.gov/BLAST/, Altschul et al., 1990). The data discussed in this work have been deposited in NCBI's Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/, Edgar et al., 2002) under GEO Series Accession no. GSE7145.
Real-time quantitative PCR (qPCR) assays
The transcript abundance of 16 genes that were differentially expressed in the macroarray experiments were selected for real-time PCR analyses. qPCR was performed with the real-time PCR system, LightCycler™ (Roche Diagnostics, Mannheim, Germany), using SYBR® Green as a fluorescent dye to measure DNA amplified products derived from the mRNA. A 100 ng aliquot of mRNA was used as a template for reverse transcription reactions to synthesize single-stranded cDNA, using MMVL-RT reverse transcriptase (Promega, Madison, WI, USA) and an oligo(dT) primer (Invitrogen, Breda, The Netherlands), according to standard procedures. Gene-specific primers were designed by using Primer Premier 5.0 software (Premier Biosoft International, Palo Alto, CA, USA) and synthesized by Alpha DNA (Montreal, Quebec). Primer sequences, annealing temperatures, and amplicon lengths are given in Table 1. For each gene, a calibration curve was performed by measuring the fluorescence of four serial dilutions (101–10−2 pg μl−1) of a plasmid bearing the fragment to amplify that served for the estimation of copy number in total cDNA.
Prunus persica primers used for the real-time PCR LightCycler™ system
| Target | Amplicon (bp) | Sequence (5′→3′) | Tm (°C) |
| Cobra (Cob) | 314 | Forward: ACT CAT CCA GGA AGC TGT GTA G | 66 |
| Reverse: ATG GCT GTA TCA TTT ATT GTC GCA | |||
| Glucan synthase | 337 | Forward: TGG GAA ACA TGG TGG TAT GAG GA | 66 |
| Reverse: CGA GAC ATT TGA AGT GAG TGA AC | |||
| Galactosyltransferase family protein | 317 | Forward: ATG TGA AAA GTG GAT GCG GAA TG | 66 |
| Reverse: TTG GAT GAG AAG CGG GAA GAG A | |||
| Endopolygalacturonase (EndoPG) | 544 | Forward: GTC ATC TGG TGT CAC AAT C | 54 |
| Reverse: ACC CTC AGT TGT TCC ATC | |||
| Cinnamoyl-CoA reductase (CCR) | 312 | Forward: ATC AAG TCC AAG ACC CCG AGA A | 66 |
| Reverse: CGC CCA ACA CGG TGC CAG GA | |||
| Pectate lyase (PL) | 294 | Forward: GCC TTG CCG TAC GCT CAT GTC | 65 |
| Reverse: CTT CAG CCT CAA CCC CTT CCC T | |||
| ER lumen protein retaining receptor 2 (ERD2) | 317 | Forward: GCC AGT ATT TTG GTC CTC CTT C | 65 |
| Reverse: TCT TGA AAT GTG AAT TCC TCG TG | |||
| Coatomer protein gamma 2-subunit (gamma 2-COP) | 350 | Forward: AAT GGG CGA CCG TCC TTT TTA C | 66 |
| Reverse: CAT CGA GCG ATC CAC ACT AGA T | |||
| Golgi transport protein SFT2-like | 268 | Forward: CAC TCA AAG GCC CGA AGA ATC A | 66 |
| Reverse: CAA CGT CAC CTC CCA AAA CAT C | |||
| SNARE-like protein Vap 27-2 | 255 | Forward: CCA AAG AAA TAC TGC GTG CGG C | 65 |
| Reverse: GGA AGA GGG TGG GCT GAT GAG | |||
| Dynamin-like protein 1A (ADL1A) | 327 | Forward: GTG AAC AAA ATC CAA AGA GCT TG | 64 |
| Reverse: GCC AGT TTC TCG ATC TGT CTC | |||
| ROC7 cyclophilin | 249 | Forward: CCA GGC AAA GAA GTC AAA GGA G | 66 |
| Reverse: TCA CCT CCC TGA ATC ATG AAA CT | |||
| Rab GTP-binding protein (Rab5) | 225 | Forward: CCA TAG GTG CTG CCT TCT TCT C | 66 |
| Reverse: CCA TGT TTG GAT TGC CTT GTG ATT | |||
| Vesicle-associated membrane protein 722 (Vamp 722) | 356 | Forward: GCA AAG CAG GTG GTC TCA GG | 64 |
| Reverse: TTA AGG CTA TTG GCA GGG GCT | |||
| Clathrin-binding protein β-adaptin | 269 | Forward: CTT GGT GAT CTG ATT GGC ATG G | 66 |
| Reverse: ACT TGT GGA ACC TGA AGG GGT C | |||
| Rab GTP-binding protein (Rab11) | 316 | Forward: ATG TTT GTA GGT TAT TAG TCG CTT A | 66 |
| Reverse: CGC TCT TGA CCA GTT GTA TCC CA |
| Target | Amplicon (bp) | Sequence (5′→3′) | Tm (°C) |
| Cobra (Cob) | 314 | Forward: ACT CAT CCA GGA AGC TGT GTA G | 66 |
| Reverse: ATG GCT GTA TCA TTT ATT GTC GCA | |||
| Glucan synthase | 337 | Forward: TGG GAA ACA TGG TGG TAT GAG GA | 66 |
| Reverse: CGA GAC ATT TGA AGT GAG TGA AC | |||
| Galactosyltransferase family protein | 317 | Forward: ATG TGA AAA GTG GAT GCG GAA TG | 66 |
| Reverse: TTG GAT GAG AAG CGG GAA GAG A | |||
| Endopolygalacturonase (EndoPG) | 544 | Forward: GTC ATC TGG TGT CAC AAT C | 54 |
| Reverse: ACC CTC AGT TGT TCC ATC | |||
| Cinnamoyl-CoA reductase (CCR) | 312 | Forward: ATC AAG TCC AAG ACC CCG AGA A | 66 |
| Reverse: CGC CCA ACA CGG TGC CAG GA | |||
| Pectate lyase (PL) | 294 | Forward: GCC TTG CCG TAC GCT CAT GTC | 65 |
| Reverse: CTT CAG CCT CAA CCC CTT CCC T | |||
| ER lumen protein retaining receptor 2 (ERD2) | 317 | Forward: GCC AGT ATT TTG GTC CTC CTT C | 65 |
| Reverse: TCT TGA AAT GTG AAT TCC TCG TG | |||
| Coatomer protein gamma 2-subunit (gamma 2-COP) | 350 | Forward: AAT GGG CGA CCG TCC TTT TTA C | 66 |
| Reverse: CAT CGA GCG ATC CAC ACT AGA T | |||
| Golgi transport protein SFT2-like | 268 | Forward: CAC TCA AAG GCC CGA AGA ATC A | 66 |
| Reverse: CAA CGT CAC CTC CCA AAA CAT C | |||
| SNARE-like protein Vap 27-2 | 255 | Forward: CCA AAG AAA TAC TGC GTG CGG C | 65 |
| Reverse: GGA AGA GGG TGG GCT GAT GAG | |||
| Dynamin-like protein 1A (ADL1A) | 327 | Forward: GTG AAC AAA ATC CAA AGA GCT TG | 64 |
| Reverse: GCC AGT TTC TCG ATC TGT CTC | |||
| ROC7 cyclophilin | 249 | Forward: CCA GGC AAA GAA GTC AAA GGA G | 66 |
| Reverse: TCA CCT CCC TGA ATC ATG AAA CT | |||
| Rab GTP-binding protein (Rab5) | 225 | Forward: CCA TAG GTG CTG CCT TCT TCT C | 66 |
| Reverse: CCA TGT TTG GAT TGC CTT GTG ATT | |||
| Vesicle-associated membrane protein 722 (Vamp 722) | 356 | Forward: GCA AAG CAG GTG GTC TCA GG | 64 |
| Reverse: TTA AGG CTA TTG GCA GGG GCT | |||
| Clathrin-binding protein β-adaptin | 269 | Forward: CTT GGT GAT CTG ATT GGC ATG G | 66 |
| Reverse: ACT TGT GGA ACC TGA AGG GGT C | |||
| Rab GTP-binding protein (Rab11) | 316 | Forward: ATG TTT GTA GGT TAT TAG TCG CTT A | 66 |
| Reverse: CGC TCT TGA CCA GTT GTA TCC CA |
Prunus persica primers used for the real-time PCR LightCycler™ system
| Target | Amplicon (bp) | Sequence (5′→3′) | Tm (°C) |
| Cobra (Cob) | 314 | Forward: ACT CAT CCA GGA AGC TGT GTA G | 66 |
| Reverse: ATG GCT GTA TCA TTT ATT GTC GCA | |||
| Glucan synthase | 337 | Forward: TGG GAA ACA TGG TGG TAT GAG GA | 66 |
| Reverse: CGA GAC ATT TGA AGT GAG TGA AC | |||
| Galactosyltransferase family protein | 317 | Forward: ATG TGA AAA GTG GAT GCG GAA TG | 66 |
| Reverse: TTG GAT GAG AAG CGG GAA GAG A | |||
| Endopolygalacturonase (EndoPG) | 544 | Forward: GTC ATC TGG TGT CAC AAT C | 54 |
| Reverse: ACC CTC AGT TGT TCC ATC | |||
| Cinnamoyl-CoA reductase (CCR) | 312 | Forward: ATC AAG TCC AAG ACC CCG AGA A | 66 |
| Reverse: CGC CCA ACA CGG TGC CAG GA | |||
| Pectate lyase (PL) | 294 | Forward: GCC TTG CCG TAC GCT CAT GTC | 65 |
| Reverse: CTT CAG CCT CAA CCC CTT CCC T | |||
| ER lumen protein retaining receptor 2 (ERD2) | 317 | Forward: GCC AGT ATT TTG GTC CTC CTT C | 65 |
| Reverse: TCT TGA AAT GTG AAT TCC TCG TG | |||
| Coatomer protein gamma 2-subunit (gamma 2-COP) | 350 | Forward: AAT GGG CGA CCG TCC TTT TTA C | 66 |
| Reverse: CAT CGA GCG ATC CAC ACT AGA T | |||
| Golgi transport protein SFT2-like | 268 | Forward: CAC TCA AAG GCC CGA AGA ATC A | 66 |
| Reverse: CAA CGT CAC CTC CCA AAA CAT C | |||
| SNARE-like protein Vap 27-2 | 255 | Forward: CCA AAG AAA TAC TGC GTG CGG C | 65 |
| Reverse: GGA AGA GGG TGG GCT GAT GAG | |||
| Dynamin-like protein 1A (ADL1A) | 327 | Forward: GTG AAC AAA ATC CAA AGA GCT TG | 64 |
| Reverse: GCC AGT TTC TCG ATC TGT CTC | |||
| ROC7 cyclophilin | 249 | Forward: CCA GGC AAA GAA GTC AAA GGA G | 66 |
| Reverse: TCA CCT CCC TGA ATC ATG AAA CT | |||
| Rab GTP-binding protein (Rab5) | 225 | Forward: CCA TAG GTG CTG CCT TCT TCT C | 66 |
| Reverse: CCA TGT TTG GAT TGC CTT GTG ATT | |||
| Vesicle-associated membrane protein 722 (Vamp 722) | 356 | Forward: GCA AAG CAG GTG GTC TCA GG | 64 |
| Reverse: TTA AGG CTA TTG GCA GGG GCT | |||
| Clathrin-binding protein β-adaptin | 269 | Forward: CTT GGT GAT CTG ATT GGC ATG G | 66 |
| Reverse: ACT TGT GGA ACC TGA AGG GGT C | |||
| Rab GTP-binding protein (Rab11) | 316 | Forward: ATG TTT GTA GGT TAT TAG TCG CTT A | 66 |
| Reverse: CGC TCT TGA CCA GTT GTA TCC CA |
| Target | Amplicon (bp) | Sequence (5′→3′) | Tm (°C) |
| Cobra (Cob) | 314 | Forward: ACT CAT CCA GGA AGC TGT GTA G | 66 |
| Reverse: ATG GCT GTA TCA TTT ATT GTC GCA | |||
| Glucan synthase | 337 | Forward: TGG GAA ACA TGG TGG TAT GAG GA | 66 |
| Reverse: CGA GAC ATT TGA AGT GAG TGA AC | |||
| Galactosyltransferase family protein | 317 | Forward: ATG TGA AAA GTG GAT GCG GAA TG | 66 |
| Reverse: TTG GAT GAG AAG CGG GAA GAG A | |||
| Endopolygalacturonase (EndoPG) | 544 | Forward: GTC ATC TGG TGT CAC AAT C | 54 |
| Reverse: ACC CTC AGT TGT TCC ATC | |||
| Cinnamoyl-CoA reductase (CCR) | 312 | Forward: ATC AAG TCC AAG ACC CCG AGA A | 66 |
| Reverse: CGC CCA ACA CGG TGC CAG GA | |||
| Pectate lyase (PL) | 294 | Forward: GCC TTG CCG TAC GCT CAT GTC | 65 |
| Reverse: CTT CAG CCT CAA CCC CTT CCC T | |||
| ER lumen protein retaining receptor 2 (ERD2) | 317 | Forward: GCC AGT ATT TTG GTC CTC CTT C | 65 |
| Reverse: TCT TGA AAT GTG AAT TCC TCG TG | |||
| Coatomer protein gamma 2-subunit (gamma 2-COP) | 350 | Forward: AAT GGG CGA CCG TCC TTT TTA C | 66 |
| Reverse: CAT CGA GCG ATC CAC ACT AGA T | |||
| Golgi transport protein SFT2-like | 268 | Forward: CAC TCA AAG GCC CGA AGA ATC A | 66 |
| Reverse: CAA CGT CAC CTC CCA AAA CAT C | |||
| SNARE-like protein Vap 27-2 | 255 | Forward: CCA AAG AAA TAC TGC GTG CGG C | 65 |
| Reverse: GGA AGA GGG TGG GCT GAT GAG | |||
| Dynamin-like protein 1A (ADL1A) | 327 | Forward: GTG AAC AAA ATC CAA AGA GCT TG | 64 |
| Reverse: GCC AGT TTC TCG ATC TGT CTC | |||
| ROC7 cyclophilin | 249 | Forward: CCA GGC AAA GAA GTC AAA GGA G | 66 |
| Reverse: TCA CCT CCC TGA ATC ATG AAA CT | |||
| Rab GTP-binding protein (Rab5) | 225 | Forward: CCA TAG GTG CTG CCT TCT TCT C | 66 |
| Reverse: CCA TGT TTG GAT TGC CTT GTG ATT | |||
| Vesicle-associated membrane protein 722 (Vamp 722) | 356 | Forward: GCA AAG CAG GTG GTC TCA GG | 64 |
| Reverse: TTA AGG CTA TTG GCA GGG GCT | |||
| Clathrin-binding protein β-adaptin | 269 | Forward: CTT GGT GAT CTG ATT GGC ATG G | 66 |
| Reverse: ACT TGT GGA ACC TGA AGG GGT C | |||
| Rab GTP-binding protein (Rab11) | 316 | Forward: ATG TTT GTA GGT TAT TAG TCG CTT A | 66 |
| Reverse: CGC TCT TGA CCA GTT GTA TCC CA |
The amplification reaction was carried out in a total volume of 20 ml containing 1 pmol of each primer, 5 mM MgCl2, 1 ml of LightCycler™ DNA Master SYBR® Green I (containing 1.25 U of Taq polymerase, 10× Taq buffer (500 mM KCl, 100 mM TRIS-HCl, pH 8.3), dNTPs each at 2 mM, 10× SYBR® Green I; Roche Diagnostics) and 100 ng of cDNA prepared as described above. The thermal cycle conditions were: denaturation at 95 °C for 10 min, followed by 35 three-step cycles of template denaturation at 95 °C with a 2 s hold, primer annealing at 60–65 °C for 15 s, and extension at 72 °C for 25 s. Fluorescence data were collected after each extension step. Melting curve analyses were performed by heating the template at 95 °C with a 0 s hold, then cooling to 65 °C with a 15 s hold, and finally increasing the temperature to 95 °C with a 0.1 °C s−1 temperature transition rate while continuously monitoring the fluorescence. All other phases were performed with a 20 °C s−1 transition rate. Fluorescence was analysed using LightCycler™ Analysis Software. The crossing point for each reaction was determined using the second derivative maximum algorithm and manual baseline adjustment. In all cases, the melting curves were checked for single peaks, and the amplification product sizes were confirmed in an agarose gel to ensure the absence of non-specific PCR products. Quadruplicate qPCR experiments were performed for each sample and the expression values were normalized against α-tubulin. To test whether α-tubulin behaves as a housekeeping gene in the analysed samples, cDNA samples from the entire set of woolly and juicy fruit analysed by qPCR were synthesized with dap spike mRNA added as internal control (0.01%). For each cDNA, transcript abundances of α-tubulin and dap were analysed by qPCR and the ratios of control transcript to the endogenous transcript α-tubulin were calculated. The results indicated that the abundance of α-tubulin mRNA remains stable between samples (data not shown). qPCR data were subjected to statistical analyses of variance and means were separated by LSD test at the 5% level of significance using Statgraphics Plus 5 (Manugistics, Inc., Rockville, MD, USA).
Results
Macroarray construction and experimental design
Peaches (P. persica cv. O'Henry) were exposed to three different treatments that simulate post-harvest conditions: (c1) 7 d at 21 °C; (c2) 15 d at 4 °C; and (c3) 15 d at 4 °C followed by 7 d at 21 °C (Campos-Vargas et al., 2006). The cold storage period of 15 d was selected to ensure the presence of woolly peaches and to reduce the possibility of obtaining fruit samples showing quality problems related to senescence that may be observed with longer storage intervals. Changes in gene expression levels between woolly and juicy fruit were analysed in three biological samples collected from juicy fruit (harvest and ripened) and woolly fruit (harvest, cold storage, and ripened). Thirty-six labelled probes corresponding to two post-harvest conditions × six experimental replications ×three biological replications were hybridized under the same conditions onto nylon macroarrays containing 847 non-redundant ESTs from a ripe peach fruit cDNA library (cv. Loring). Gene expression values were measured as described in the Materials and methods. A high correlation coefficient between the signal levels was found across the different experiments (mean >0.84, data not shown), indicating an important degree of similarity in expression profiles. The quantification data files generated from the macroarray analyses were processed to remove low-quality spots and normalized as described in the Materials and methods. The results of these analysis showed that 68% of the clones (n=573) fulfilled all filter criteria and they were selected for sequence analysis (a full listing of clones is available in Supplementary Data of the Supplementary data available at JXB online). Seventy-five cDNAs clones were differentially expressed at an FDR <10% between woolly and juicy peaches as determined by SAM (Tusher et al., 2001), and 31 genes were detected in only one condition while in the other a value was assigned to them as described in Materials and methods. The majority of these genes (n=97) were down-regulated in the woolly fruit, suggesting that the main part of the fruit response to a post-harvest condition that leads to woolliness involves gene down-regulation. For a better interpretation of the results, these regulated genes were classified in 13 functional categories according to their putative role in cell metabolism taking into account functions reported for gene homologues in other systems (see Supplementary Data of the Supplementary data at JXB online). Most of the differentially expressed genes showed a high sequence homology with other species, mainly Arabidopsis thaliana and Oryza sativa, with >40% sequence identity at the amino acid level. The sequence identities and percentage of P. persica sequence length that aligned with the corresponding homologue (Cob >80%) suggests a high degree of conservation of these gene products in plants. Genes of particular biological interest, cell wall and endomembrane trafficking, were selected for qPCR validation and further discussion of their potential role in peach woolliness. They are described in Table 2 and discussed below.
Selected genes differentially expressed between juicy and woolly fruit
| Clone IDa | Homologue definition | Organism | GenBank | COBb | E value | IDENTc | SIMd | CAZye | Experimental conditionf | |
| J | W | |||||||||
| Genes with some function related to cell wall metabolism | ||||||||||
| PP_LEa0003N12f | Cobra (Cob) | A. thaliana | NP_568930 | 96 | 0.0 | 78 | 88 | – | 1.80 | 0.56 |
| PP_LEa0004K19f | Glucan synthase | O. sativa | NP_916159 | 100 | 7e-75 | 68 | 85 | GT48 | 1.25 | 0.80 |
| PP_LEa0005N23f | Galactosyltransferase family protein | A. thaliana | NP_193838 | 99 | 2e-80 | 72 | 86 | GT31 | 1.84 | 0.54 |
| PP_LEa0006P14f | Endopolygalacturonase (EndoPG) | P. persica | AAC64184 | 100 | 0.0 | 99 | 100 | GH28 | 3.23 | 0.31 |
| PP_LEa0008K15f | Cinnamoyl-CoA reductase (CCR) | A. thaliana | NP_200657 | 89 | 1e-81 | 75 | 88 | – | 1.99 | 0.50 |
| PP_LEa0015N02f | Pectate lyase (PL) | A. thaliana | NP_191074 | 100 | 8e-90 | 80 | 92 | PL1 | 1.93 | 0.52 |
| PP_LEa0033B07f | Cotton fibre expressed protein 2 | G. hirsutum | AAC33277 | 95 | 2e-18 | 40 | 55 | – | 2.17 | 0.46 |
| Genes related to endomembrane traffic | ||||||||||
| PP_LEa0001H21f | ER lumen protein retaining receptor 2 (ERD2) | A. thaliana | NP_564326 | 99 | 2e-72 | 75 | 90 | n/a | 1.85 | 0.54 |
| PP_LEa0004B09f | Coatomer protein gamma 2-subunit (gamma 2-COP) | O. sativa | BAC84213 | 90 | 9e-160 | 89 | 95 | n/a | 0.85 | 1.17 |
| PP_LEa0008F22f | Golgi transport protein SFT2-like | A. thaliana | NP_567749 | 100 | 5e-96 | 78 | 87 | n/a | 1.45 | 0.68 |
| PP_LEa0009A08f | SNARE-like protein Vap 27–2 | A. thaliana | NP_172359 | 93 | 7e-46 | 41 | 53 | n/a | 1.24 | 0.81 |
| PP_LEa0009I13f | Dynamin-like protein 1A (ADL1A) | G. max | AAB05992 | 99 | 4e-94 | 94 | 97 | n/a | 1.34 | 0.74 |
| PP_LEa0009K22f | ROC7 cyclophilin | A. thaliana | NP_200679 | 100 | 7e-89 | 82 | 92 | n/a | 1.74 | 0.57 |
| PP_LEa0013L18f | Rab GTP-binding protein (Rab5) | N. tabacum | CAA50609 | 96 | 5e-79 | 89 | 96 | n/a | 1.89 | 0.53 |
| PP_LEa0014G24f | Vesicle-associated membrane protein 722 (Vamp 722) | O. sativa | CAD70274 | 99 | 5e-99 | 86 | 94 | n/a | 2.10 | 0.48 |
| PP_LEa0017D15f | Clathrin-binding protein β-adaptin | A. thaliana | NP_192877 | 89 | 2e-35 | 75 | 88 | n/a | 0.75 | 1.33 |
| PP_LEa0020B02f | Rab GTP-binding protein (Rab11) | G. hirsutum | AAD48018 | 80 | 1e-38 | 85 | 92 | n/a | 1.56 | 0.64 |
| PP_LEa0024O23f | GTPase-activating protein Sec23 | A. thaliana | NP_563741 | 86 | 1e-91 | 89 | 94 | n/a | 2.77 | 0.36 |
| PP_LEa0030L15f | Secretory carrier membrane protein like (SCAMP-like) | P. sativum | AAC82326 | 96 | 9e-125 | 75 | 88 | n/a | 1.52 | 0.66 |
| Clone IDa | Homologue definition | Organism | GenBank | COBb | E value | IDENTc | SIMd | CAZye | Experimental conditionf | |
| J | W | |||||||||
| Genes with some function related to cell wall metabolism | ||||||||||
| PP_LEa0003N12f | Cobra (Cob) | A. thaliana | NP_568930 | 96 | 0.0 | 78 | 88 | – | 1.80 | 0.56 |
| PP_LEa0004K19f | Glucan synthase | O. sativa | NP_916159 | 100 | 7e-75 | 68 | 85 | GT48 | 1.25 | 0.80 |
| PP_LEa0005N23f | Galactosyltransferase family protein | A. thaliana | NP_193838 | 99 | 2e-80 | 72 | 86 | GT31 | 1.84 | 0.54 |
| PP_LEa0006P14f | Endopolygalacturonase (EndoPG) | P. persica | AAC64184 | 100 | 0.0 | 99 | 100 | GH28 | 3.23 | 0.31 |
| PP_LEa0008K15f | Cinnamoyl-CoA reductase (CCR) | A. thaliana | NP_200657 | 89 | 1e-81 | 75 | 88 | – | 1.99 | 0.50 |
| PP_LEa0015N02f | Pectate lyase (PL) | A. thaliana | NP_191074 | 100 | 8e-90 | 80 | 92 | PL1 | 1.93 | 0.52 |
| PP_LEa0033B07f | Cotton fibre expressed protein 2 | G. hirsutum | AAC33277 | 95 | 2e-18 | 40 | 55 | – | 2.17 | 0.46 |
| Genes related to endomembrane traffic | ||||||||||
| PP_LEa0001H21f | ER lumen protein retaining receptor 2 (ERD2) | A. thaliana | NP_564326 | 99 | 2e-72 | 75 | 90 | n/a | 1.85 | 0.54 |
| PP_LEa0004B09f | Coatomer protein gamma 2-subunit (gamma 2-COP) | O. sativa | BAC84213 | 90 | 9e-160 | 89 | 95 | n/a | 0.85 | 1.17 |
| PP_LEa0008F22f | Golgi transport protein SFT2-like | A. thaliana | NP_567749 | 100 | 5e-96 | 78 | 87 | n/a | 1.45 | 0.68 |
| PP_LEa0009A08f | SNARE-like protein Vap 27–2 | A. thaliana | NP_172359 | 93 | 7e-46 | 41 | 53 | n/a | 1.24 | 0.81 |
| PP_LEa0009I13f | Dynamin-like protein 1A (ADL1A) | G. max | AAB05992 | 99 | 4e-94 | 94 | 97 | n/a | 1.34 | 0.74 |
| PP_LEa0009K22f | ROC7 cyclophilin | A. thaliana | NP_200679 | 100 | 7e-89 | 82 | 92 | n/a | 1.74 | 0.57 |
| PP_LEa0013L18f | Rab GTP-binding protein (Rab5) | N. tabacum | CAA50609 | 96 | 5e-79 | 89 | 96 | n/a | 1.89 | 0.53 |
| PP_LEa0014G24f | Vesicle-associated membrane protein 722 (Vamp 722) | O. sativa | CAD70274 | 99 | 5e-99 | 86 | 94 | n/a | 2.10 | 0.48 |
| PP_LEa0017D15f | Clathrin-binding protein β-adaptin | A. thaliana | NP_192877 | 89 | 2e-35 | 75 | 88 | n/a | 0.75 | 1.33 |
| PP_LEa0020B02f | Rab GTP-binding protein (Rab11) | G. hirsutum | AAD48018 | 80 | 1e-38 | 85 | 92 | n/a | 1.56 | 0.64 |
| PP_LEa0024O23f | GTPase-activating protein Sec23 | A. thaliana | NP_563741 | 86 | 1e-91 | 89 | 94 | n/a | 2.77 | 0.36 |
| PP_LEa0030L15f | Secretory carrier membrane protein like (SCAMP-like) | P. sativum | AAC82326 | 96 | 9e-125 | 75 | 88 | n/a | 1.52 | 0.66 |
Nineteen differentially expressed genes were grouped into two physiological processes: cell wall metabolism and endomembrane traffic.
Clone identification according to Clemson University Genomics Institute database (http://www.genome.clemson.edu/projects/peach/est/).
COB, percentage of P. persica sequence length that aligned with the corresponding homologue.
IDENT and
SIM, the identity and similarity percentages, were obtained using the BLOSUM62 matrix (http://www.ncbi.nlm.nih.gov).
CAZy database classification (http://afmb.cnrs-mrs.fr/∼pedro/CAZY).
Expression ratios of juicy/woolly (J) and woolly/juicy (W).
Selected genes differentially expressed between juicy and woolly fruit
| Clone IDa | Homologue definition | Organism | GenBank | COBb | E value | IDENTc | SIMd | CAZye | Experimental conditionf | |
| J | W | |||||||||
| Genes with some function related to cell wall metabolism | ||||||||||
| PP_LEa0003N12f | Cobra (Cob) | A. thaliana | NP_568930 | 96 | 0.0 | 78 | 88 | – | 1.80 | 0.56 |
| PP_LEa0004K19f | Glucan synthase | O. sativa | NP_916159 | 100 | 7e-75 | 68 | 85 | GT48 | 1.25 | 0.80 |
| PP_LEa0005N23f | Galactosyltransferase family protein | A. thaliana | NP_193838 | 99 | 2e-80 | 72 | 86 | GT31 | 1.84 | 0.54 |
| PP_LEa0006P14f | Endopolygalacturonase (EndoPG) | P. persica | AAC64184 | 100 | 0.0 | 99 | 100 | GH28 | 3.23 | 0.31 |
| PP_LEa0008K15f | Cinnamoyl-CoA reductase (CCR) | A. thaliana | NP_200657 | 89 | 1e-81 | 75 | 88 | – | 1.99 | 0.50 |
| PP_LEa0015N02f | Pectate lyase (PL) | A. thaliana | NP_191074 | 100 | 8e-90 | 80 | 92 | PL1 | 1.93 | 0.52 |
| PP_LEa0033B07f | Cotton fibre expressed protein 2 | G. hirsutum | AAC33277 | 95 | 2e-18 | 40 | 55 | – | 2.17 | 0.46 |
| Genes related to endomembrane traffic | ||||||||||
| PP_LEa0001H21f | ER lumen protein retaining receptor 2 (ERD2) | A. thaliana | NP_564326 | 99 | 2e-72 | 75 | 90 | n/a | 1.85 | 0.54 |
| PP_LEa0004B09f | Coatomer protein gamma 2-subunit (gamma 2-COP) | O. sativa | BAC84213 | 90 | 9e-160 | 89 | 95 | n/a | 0.85 | 1.17 |
| PP_LEa0008F22f | Golgi transport protein SFT2-like | A. thaliana | NP_567749 | 100 | 5e-96 | 78 | 87 | n/a | 1.45 | 0.68 |
| PP_LEa0009A08f | SNARE-like protein Vap 27–2 | A. thaliana | NP_172359 | 93 | 7e-46 | 41 | 53 | n/a | 1.24 | 0.81 |
| PP_LEa0009I13f | Dynamin-like protein 1A (ADL1A) | G. max | AAB05992 | 99 | 4e-94 | 94 | 97 | n/a | 1.34 | 0.74 |
| PP_LEa0009K22f | ROC7 cyclophilin | A. thaliana | NP_200679 | 100 | 7e-89 | 82 | 92 | n/a | 1.74 | 0.57 |
| PP_LEa0013L18f | Rab GTP-binding protein (Rab5) | N. tabacum | CAA50609 | 96 | 5e-79 | 89 | 96 | n/a | 1.89 | 0.53 |
| PP_LEa0014G24f | Vesicle-associated membrane protein 722 (Vamp 722) | O. sativa | CAD70274 | 99 | 5e-99 | 86 | 94 | n/a | 2.10 | 0.48 |
| PP_LEa0017D15f | Clathrin-binding protein β-adaptin | A. thaliana | NP_192877 | 89 | 2e-35 | 75 | 88 | n/a | 0.75 | 1.33 |
| PP_LEa0020B02f | Rab GTP-binding protein (Rab11) | G. hirsutum | AAD48018 | 80 | 1e-38 | 85 | 92 | n/a | 1.56 | 0.64 |
| PP_LEa0024O23f | GTPase-activating protein Sec23 | A. thaliana | NP_563741 | 86 | 1e-91 | 89 | 94 | n/a | 2.77 | 0.36 |
| PP_LEa0030L15f | Secretory carrier membrane protein like (SCAMP-like) | P. sativum | AAC82326 | 96 | 9e-125 | 75 | 88 | n/a | 1.52 | 0.66 |
| Clone IDa | Homologue definition | Organism | GenBank | COBb | E value | IDENTc | SIMd | CAZye | Experimental conditionf | |
| J | W | |||||||||
| Genes with some function related to cell wall metabolism | ||||||||||
| PP_LEa0003N12f | Cobra (Cob) | A. thaliana | NP_568930 | 96 | 0.0 | 78 | 88 | – | 1.80 | 0.56 |
| PP_LEa0004K19f | Glucan synthase | O. sativa | NP_916159 | 100 | 7e-75 | 68 | 85 | GT48 | 1.25 | 0.80 |
| PP_LEa0005N23f | Galactosyltransferase family protein | A. thaliana | NP_193838 | 99 | 2e-80 | 72 | 86 | GT31 | 1.84 | 0.54 |
| PP_LEa0006P14f | Endopolygalacturonase (EndoPG) | P. persica | AAC64184 | 100 | 0.0 | 99 | 100 | GH28 | 3.23 | 0.31 |
| PP_LEa0008K15f | Cinnamoyl-CoA reductase (CCR) | A. thaliana | NP_200657 | 89 | 1e-81 | 75 | 88 | – | 1.99 | 0.50 |
| PP_LEa0015N02f | Pectate lyase (PL) | A. thaliana | NP_191074 | 100 | 8e-90 | 80 | 92 | PL1 | 1.93 | 0.52 |
| PP_LEa0033B07f | Cotton fibre expressed protein 2 | G. hirsutum | AAC33277 | 95 | 2e-18 | 40 | 55 | – | 2.17 | 0.46 |
| Genes related to endomembrane traffic | ||||||||||
| PP_LEa0001H21f | ER lumen protein retaining receptor 2 (ERD2) | A. thaliana | NP_564326 | 99 | 2e-72 | 75 | 90 | n/a | 1.85 | 0.54 |
| PP_LEa0004B09f | Coatomer protein gamma 2-subunit (gamma 2-COP) | O. sativa | BAC84213 | 90 | 9e-160 | 89 | 95 | n/a | 0.85 | 1.17 |
| PP_LEa0008F22f | Golgi transport protein SFT2-like | A. thaliana | NP_567749 | 100 | 5e-96 | 78 | 87 | n/a | 1.45 | 0.68 |
| PP_LEa0009A08f | SNARE-like protein Vap 27–2 | A. thaliana | NP_172359 | 93 | 7e-46 | 41 | 53 | n/a | 1.24 | 0.81 |
| PP_LEa0009I13f | Dynamin-like protein 1A (ADL1A) | G. max | AAB05992 | 99 | 4e-94 | 94 | 97 | n/a | 1.34 | 0.74 |
| PP_LEa0009K22f | ROC7 cyclophilin | A. thaliana | NP_200679 | 100 | 7e-89 | 82 | 92 | n/a | 1.74 | 0.57 |
| PP_LEa0013L18f | Rab GTP-binding protein (Rab5) | N. tabacum | CAA50609 | 96 | 5e-79 | 89 | 96 | n/a | 1.89 | 0.53 |
| PP_LEa0014G24f | Vesicle-associated membrane protein 722 (Vamp 722) | O. sativa | CAD70274 | 99 | 5e-99 | 86 | 94 | n/a | 2.10 | 0.48 |
| PP_LEa0017D15f | Clathrin-binding protein β-adaptin | A. thaliana | NP_192877 | 89 | 2e-35 | 75 | 88 | n/a | 0.75 | 1.33 |
| PP_LEa0020B02f | Rab GTP-binding protein (Rab11) | G. hirsutum | AAD48018 | 80 | 1e-38 | 85 | 92 | n/a | 1.56 | 0.64 |
| PP_LEa0024O23f | GTPase-activating protein Sec23 | A. thaliana | NP_563741 | 86 | 1e-91 | 89 | 94 | n/a | 2.77 | 0.36 |
| PP_LEa0030L15f | Secretory carrier membrane protein like (SCAMP-like) | P. sativum | AAC82326 | 96 | 9e-125 | 75 | 88 | n/a | 1.52 | 0.66 |
Nineteen differentially expressed genes were grouped into two physiological processes: cell wall metabolism and endomembrane traffic.
Clone identification according to Clemson University Genomics Institute database (http://www.genome.clemson.edu/projects/peach/est/).
COB, percentage of P. persica sequence length that aligned with the corresponding homologue.
IDENT and
SIM, the identity and similarity percentages, were obtained using the BLOSUM62 matrix (http://www.ncbi.nlm.nih.gov).
CAZy database classification (http://afmb.cnrs-mrs.fr/∼pedro/CAZY).
Expression ratios of juicy/woolly (J) and woolly/juicy (W).
Confirmation of gene expression patterns by real-time qPCR
In order to validate macroarray data, the relative transcript abundance of 16 differentially expressed genes was tested by qPCR, using cDNA from juicy and woolly samples as templates. Genes were selected based on their involvement in a cellular process that may have an influence on fruit woolliness. All reactions were performed in quadruplicate to determine significant differences in transcript abundance between fruit samples (t-test, P < 0.05). For each qPCR, gene expression was normalized to a reference mRNA (see Materials and methods). The results of expression data obtained by qPCR were in agreement (up- or down-regulation) with those obtained by macroarray analysis (Table 3). Although, in most of the cases, the magnitude of gene expression differences was similar between macroarray and qPCR assays, five out of the 16 differentially expressed genes confirmed by real-time PCR showed expression differences greater than that determined by cDNA macroarray, a phenomenon that has been observed before (Rajeevan et al., 2001; Yuen et al., 2002). This might be due to cross-hybridization between related sequences. The cDNA clones on the array might contain domains with high sequence identity between isoforms, which would lead to overestimation of the signal intensity obtained from transcripts present in the untreated samples. However, the authors are confident that the qPCR assays specifically amplified each cDNA with no cross-reactivity between isoforms.
Comparison of gene expression levels obtained by cDNA macroarray and qPCR analysis for 16 differentially expressed genes
| Clone ID | Homologue definition (GenBank accession no.)a | Fold change | |
| Macroarrayb | Real-time PCRc | ||
| PP_LEa0003N12f | Cobra (Cob) (NP_568930) | 1.80 | 3.38±0.26 |
| PP_LEa0004K19f | Glucan synthase (NP_916159) | 1.25 | 2.91±0.65 |
| PP_LEa0005N23f | Galactosyltransferase family protein (NP_193838) | 1.84 | 3.82±0.77 |
| PP_LEa0006P14f | Endopolygalacturonase (EndoPG) (AAC64184) | 3.23 | 2.50±0.69 |
| PP_LEa0008K15f | Cinnamoyl-CoA reductase (CCR) (NP_200657) | 1.99 | 4.01±1.52 |
| PP_LEa0015N02f | Pectate lyase (PL) (NP_191074) | 1.93 | 4.35±0.93 |
| PP_LEa0001H21f | ER lumen protein retaining receptor 2 (ERD2) (NP_564326) | 1.85 | 4.00±1.35 |
| PP_LEa0004B09f | Coatomer protein gamma 2-subunit (gamma 2-COP) (BAC84213) | 0.85 | –1.34±0.30 |
| PP_LEa0008F22f | Golgi transport protein SFT2-like (NP_567749) | 1.45 | 1.79±0.34 |
| PP_LEa0009A08f | SNARE-like protein Vap 27-2 (NP_172359) | 1.24 | 2.48±0.87 |
| PP_LEa0009I13f | Dynamin-like protein 1A (ADL1A) (AAB05992) | 1.34 | 6.04±0.82 |
| PP_LEa0009K22f | ROC7 cyclophilin (NP_200679) | 1.74 | 15.67±3.58 |
| PP_LEa0013L18f | Rab GTP-binding protein (Rab5) (CAA50609) | 1.89 | 2.54±0.88 |
| PP_LEa0014G24f | Vesicle-associated membrane protein 722 (Vamp 722) (CAD70274) | 2.10 | 1.28±0.49 |
| PP_LEa0017D15f | Clathrin-binding protein β-adaptin (NP_192877) | 0.75 | –1.77±0.04 |
| PP_LEa0020B02f | Rab GTP-binding protein (Rab11) (AAD48018) | 1.56 | 2.32±0.51 |
| Clone ID | Homologue definition (GenBank accession no.)a | Fold change | |
| Macroarrayb | Real-time PCRc | ||
| PP_LEa0003N12f | Cobra (Cob) (NP_568930) | 1.80 | 3.38±0.26 |
| PP_LEa0004K19f | Glucan synthase (NP_916159) | 1.25 | 2.91±0.65 |
| PP_LEa0005N23f | Galactosyltransferase family protein (NP_193838) | 1.84 | 3.82±0.77 |
| PP_LEa0006P14f | Endopolygalacturonase (EndoPG) (AAC64184) | 3.23 | 2.50±0.69 |
| PP_LEa0008K15f | Cinnamoyl-CoA reductase (CCR) (NP_200657) | 1.99 | 4.01±1.52 |
| PP_LEa0015N02f | Pectate lyase (PL) (NP_191074) | 1.93 | 4.35±0.93 |
| PP_LEa0001H21f | ER lumen protein retaining receptor 2 (ERD2) (NP_564326) | 1.85 | 4.00±1.35 |
| PP_LEa0004B09f | Coatomer protein gamma 2-subunit (gamma 2-COP) (BAC84213) | 0.85 | –1.34±0.30 |
| PP_LEa0008F22f | Golgi transport protein SFT2-like (NP_567749) | 1.45 | 1.79±0.34 |
| PP_LEa0009A08f | SNARE-like protein Vap 27-2 (NP_172359) | 1.24 | 2.48±0.87 |
| PP_LEa0009I13f | Dynamin-like protein 1A (ADL1A) (AAB05992) | 1.34 | 6.04±0.82 |
| PP_LEa0009K22f | ROC7 cyclophilin (NP_200679) | 1.74 | 15.67±3.58 |
| PP_LEa0013L18f | Rab GTP-binding protein (Rab5) (CAA50609) | 1.89 | 2.54±0.88 |
| PP_LEa0014G24f | Vesicle-associated membrane protein 722 (Vamp 722) (CAD70274) | 2.10 | 1.28±0.49 |
| PP_LEa0017D15f | Clathrin-binding protein β-adaptin (NP_192877) | 0.75 | –1.77±0.04 |
| PP_LEa0020B02f | Rab GTP-binding protein (Rab11) (AAD48018) | 1.56 | 2.32±0.51 |
Sixteen cDNA clones belonging to two functional categories, cell wall metabolism and endomembrane trafficking, were analysed by qPCR to confirm the cDNA macroarray results.
Definition and accession number of the best match according to BLASTP against the non-redundant database at NCBI.
Fold-value changes derived from macroarray analysis represent differences between normalized expression values.
The expression levels of the selected genes derived from qPCR reactions were normalized using P. persica α-tubulin.
Comparison of gene expression levels obtained by cDNA macroarray and qPCR analysis for 16 differentially expressed genes
| Clone ID | Homologue definition (GenBank accession no.)a | Fold change | |
| Macroarrayb | Real-time PCRc | ||
| PP_LEa0003N12f | Cobra (Cob) (NP_568930) | 1.80 | 3.38±0.26 |
| PP_LEa0004K19f | Glucan synthase (NP_916159) | 1.25 | 2.91±0.65 |
| PP_LEa0005N23f | Galactosyltransferase family protein (NP_193838) | 1.84 | 3.82±0.77 |
| PP_LEa0006P14f | Endopolygalacturonase (EndoPG) (AAC64184) | 3.23 | 2.50±0.69 |
| PP_LEa0008K15f | Cinnamoyl-CoA reductase (CCR) (NP_200657) | 1.99 | 4.01±1.52 |
| PP_LEa0015N02f | Pectate lyase (PL) (NP_191074) | 1.93 | 4.35±0.93 |
| PP_LEa0001H21f | ER lumen protein retaining receptor 2 (ERD2) (NP_564326) | 1.85 | 4.00±1.35 |
| PP_LEa0004B09f | Coatomer protein gamma 2-subunit (gamma 2-COP) (BAC84213) | 0.85 | –1.34±0.30 |
| PP_LEa0008F22f | Golgi transport protein SFT2-like (NP_567749) | 1.45 | 1.79±0.34 |
| PP_LEa0009A08f | SNARE-like protein Vap 27-2 (NP_172359) | 1.24 | 2.48±0.87 |
| PP_LEa0009I13f | Dynamin-like protein 1A (ADL1A) (AAB05992) | 1.34 | 6.04±0.82 |
| PP_LEa0009K22f | ROC7 cyclophilin (NP_200679) | 1.74 | 15.67±3.58 |
| PP_LEa0013L18f | Rab GTP-binding protein (Rab5) (CAA50609) | 1.89 | 2.54±0.88 |
| PP_LEa0014G24f | Vesicle-associated membrane protein 722 (Vamp 722) (CAD70274) | 2.10 | 1.28±0.49 |
| PP_LEa0017D15f | Clathrin-binding protein β-adaptin (NP_192877) | 0.75 | –1.77±0.04 |
| PP_LEa0020B02f | Rab GTP-binding protein (Rab11) (AAD48018) | 1.56 | 2.32±0.51 |
| Clone ID | Homologue definition (GenBank accession no.)a | Fold change | |
| Macroarrayb | Real-time PCRc | ||
| PP_LEa0003N12f | Cobra (Cob) (NP_568930) | 1.80 | 3.38±0.26 |
| PP_LEa0004K19f | Glucan synthase (NP_916159) | 1.25 | 2.91±0.65 |
| PP_LEa0005N23f | Galactosyltransferase family protein (NP_193838) | 1.84 | 3.82±0.77 |
| PP_LEa0006P14f | Endopolygalacturonase (EndoPG) (AAC64184) | 3.23 | 2.50±0.69 |
| PP_LEa0008K15f | Cinnamoyl-CoA reductase (CCR) (NP_200657) | 1.99 | 4.01±1.52 |
| PP_LEa0015N02f | Pectate lyase (PL) (NP_191074) | 1.93 | 4.35±0.93 |
| PP_LEa0001H21f | ER lumen protein retaining receptor 2 (ERD2) (NP_564326) | 1.85 | 4.00±1.35 |
| PP_LEa0004B09f | Coatomer protein gamma 2-subunit (gamma 2-COP) (BAC84213) | 0.85 | –1.34±0.30 |
| PP_LEa0008F22f | Golgi transport protein SFT2-like (NP_567749) | 1.45 | 1.79±0.34 |
| PP_LEa0009A08f | SNARE-like protein Vap 27-2 (NP_172359) | 1.24 | 2.48±0.87 |
| PP_LEa0009I13f | Dynamin-like protein 1A (ADL1A) (AAB05992) | 1.34 | 6.04±0.82 |
| PP_LEa0009K22f | ROC7 cyclophilin (NP_200679) | 1.74 | 15.67±3.58 |
| PP_LEa0013L18f | Rab GTP-binding protein (Rab5) (CAA50609) | 1.89 | 2.54±0.88 |
| PP_LEa0014G24f | Vesicle-associated membrane protein 722 (Vamp 722) (CAD70274) | 2.10 | 1.28±0.49 |
| PP_LEa0017D15f | Clathrin-binding protein β-adaptin (NP_192877) | 0.75 | –1.77±0.04 |
| PP_LEa0020B02f | Rab GTP-binding protein (Rab11) (AAD48018) | 1.56 | 2.32±0.51 |
Sixteen cDNA clones belonging to two functional categories, cell wall metabolism and endomembrane trafficking, were analysed by qPCR to confirm the cDNA macroarray results.
Definition and accession number of the best match according to BLASTP against the non-redundant database at NCBI.
Fold-value changes derived from macroarray analysis represent differences between normalized expression values.
The expression levels of the selected genes derived from qPCR reactions were normalized using P. persica α-tubulin.
Gene expression during cold storage and in fruit from different cultivars
In order to gain further insights into the mechanisms underlying peach woolliness, the expression levels of four genes: cobra (cob), endopolygalacturonase (endoPG), cinnamoyl-CoA-reductase (CCR), and rab11 in fruit after cold storage were determined. Peaches (cv. O'Henry) were subjected to different simulated post-harvest conditions to obtain: ripened and juicy fruit (c1); cold-stored, non-ripened fruit (c2); and cold-stored, ripened, and woolly fruit (c3), and cDNA from three fruit per treatment was used as template in duplicate qPCRs. The results (Fig. 1) indicated that the expression levels of these genes were lower in the cold-stored fruit (c2) than in fruit obtained from c1, and they remained low in the woolly fruit after ripening (c3). Further analyses that include the comparison of gene expression changes between fruit at harvest and subjected to the 21 °C and 4 °C treatments for the same period of time will be necessary to elucidate whether the detected variation in the expression patterns of these genes might be responsible for the enhanced susceptibility of peach fruit to woolliness.
Gene expression analyses after post-harvest treatments and in fruit from different cultivars. Four genes (cob, endoPG, CCR, and rab11) whose expression levels were decreased in woolly fruit, according to the macroarray data, were assayed by qPCR using cDNAs from different post-harvest treatments and cultivars. For peaches cv. O'Henry (O), the following samples were analysed: (c1) ripened and juicy fruit; (c2) cold-stored, non-ripened fruit; and (c3) cold-stored, ripened, and woolly. For peaches cv. Elegant Lady (E) and cv. Flamekist (F), samples from: (c1) ripened and juicy fruit and (c3) cold-stored, ripened, and woolly fruit were assayed. For each gene, the relative abundance of mRNA was normalized towards tubulin in the corresponding samples. The results are presented as a percentage of the highest value of relative abundance. Different letters between each treatment represent significant differences at P < 0.05 by LSD test.
Since one of the key components that modulates susceptibility to the woolliness disorder is fruit cultivar (Kader, 1985; von Mollendorf, 1987), an assessment was made of whether the gene expression changes that were observed in cv. O'Henry (O) can also be detected in woolly peaches from cv. Elegant Lady (E) and cv. Flamekist (F) (Fig. 1). The fruit from these cultivars were obtained during the same growing season and subjected to the simulated post-harvest conditions described in the Materials and methods. After measuring fruit free juice content, differences in the incidence of woolliness between these cultivars were determined, with 90% woolly fruit in O'Henry, 55% in Flamekist, and 5% in Elegant Lady peaches. qPCR was then used to compare the expression levels of cob, endoPG, CCR, and rab11 genes between juicy and woolly fruit from the different cultivars. The results (Fig. 1) indicate that the expression levels of the four tested genes were consistently lower in the samples obtained from woolly peaches when compared with the juicy fruit.
Discussion
In order to assess changes in gene expression associated with woolliness development in peaches cv. O'Henry, the expression patterns of 847 unigenes in fruit subjected to two different treatments that simulate post-harvest conditions were analysed. The results revealed that 106 genes change their relative expression levels in woolly fruit when compared with juicy fruit. The abundance of most of the transcripts (93%) decreased in woolly fruit; it is possible that this change may be the fruit response to prolonged cold storage that preceded peach ripening, a condition that commercial peaches confront during shipping to distant markets. Similarly, extensive down-regulation of gene expression (90%) has been reported for sunflower plants growing under low temperatures (Hewezi et al., 2006). In that work, the non-induction of genes involved in cold tolerance was proposed to be responsible for the sensitivity of sunflower to low temperatures.
When the transcript abundance of four genes in peaches subjected to three different post-harvest treatments was analysed, it was found that compared with the juicy fruit (c1), cold-stored (c2) and woolly fruit (c3) exhibited a significant decrease in the expression level of the selected genes. In this experiment, it was found that the expression levels of cob, endoPG, CCR, and rab11 were not significantly different between the cold-stored and the woolly fruit. In addition, qPCR analyses of samples from different peach cultivars showed that the transcript abundance of these four genes was consistently lower in woolly peaches when compared with the juicy fruit. Overall these results provide an initial characterization of the transcriptome activity of peach as a woolliness-susceptible species under different post-harvest conditions.
From a functional categorization analysis of peach cDNAs followed by qPCR assays, it was demonstrated that in the woolly fruit there is a differential expression of genes putatively involved in cell wall metabolism and endomembrane trafficking. Normal ripening of peach fruit involves a series of cell wall modifications, including changes in pectin metabolism, brought about by the secretion of a number of enzymes (see Lurie and Crisosto, 2005), and possibly deposition of newly synthesized cell proteins (Trainotti et al., 2003, 2006) that are expected to be closely correlated with the functionality of the endomembrane system.
Genes with functions in cell wall synthesis and remodelling
The results of macroarray and qPCR experiments were in general consistent with previous information and provided new data on genes potentially involved in modulating cell wall structure (Tables 2, 3). Woolliness in peaches is believed to be caused by an altered activity of cell wall enzymes, mainly EndoPG and PME, during cold storage, which affects the metabolism of cell wall polysaccharides (reviewed in Lurie and Crisosto, 2005) and leads to an imbalance in pectin degradation. It has been reported that during peach ripening the expression of Pp-endoPG is first detected at the onset of the climacteric period, reaching its maximum at later stages of softening (Trainotti et al., 2003, 2006). In the present macroarray experiments, the relative expression level of endoPG (Pp-endoPG, clone PP_LEa0006P14f) was decreased 3.2-fold in woolly fruit. The lower level of Pp-endoPG expression detected in the woolly fruit is consistent with the low ethylene evolution that accompanies the development of peach woolliness, since it has been shown that Pp-endoPG can be directly regulated by ethylene (Hayama et al., 2006).
An increase in PME activity has been reported between juicy and woolly O'Henry peaches after cold storage for 2 weeks followed by ripening at 21 °C (Brummel et al., 2004), indicating that a high PME activity correlates well with the development of woolliness. However, the macroarray hybridization analysis indicated that the expression level of five putative PME genes (see Supplementary Supplementary Data at JXB online) did not differ significantly between woolly and juicy peaches (expression data on GEO accession no. GSE7145), suggesting that post-transcriptional regulatory mechanisms might be acting to modify PME activity. In this regard, specific PME proteinaceous inhibitors, named as PMEIs (Di Matteo et al., 2005), which were discovered in kiwi (Giovane et al., 1995) and were also found in A. thaliana (Raiola et al., 2004), are able specifically to inhibit the activity of PME (Giovane et al., 2004). Therefore, the question of whether changes in the expression levels of P. persica PMEIs might increase or decrease the activity of PME under different post-harvest conditions remains open, and certainly deserves further investigation.
The expression level of a gene encoding a putative pectate lyase (PL) was reduced in woolly peaches (clone PP_LEa0015N02f). PL activity and PL genes have been detected in many higher plants where they are usually encoded by large gene families (Marin-Rodriguez et al., 2002), including sequences specifically expressed in fruit (Medina-Escobar et al., 1997). Trainotti et al. (2003) showed that the expression of two PL genes increases before the climacteric rise in ethylene production, and suggested that PL acts during early degradation of cell wall pectins, making them susceptible to the attack of endoPG and PME enzymes, which are active at later stages of development. In addition, Hayama et al. (2006) showed that the expression levels of two PL genes remained unchanged during ripening of stony hard flesh and melting-flesh peach fruit. Here, a differential expression is reported between juice and woolly peach of a putative novel PL gene (encoded by PP_LEa0015N02f) for which additional analysis of the gene expression pattern and enzymatic activity during peach ripening and under different post-harvest conditions will be necessary to determine whether the lower transcript abundance of this PL gene might contribute to the development of peach woolliness.
Three cDNAs encoding products with potential roles in the synthesis of cell wall components were identified as being down-regulated in the woolly peach (Tables 2, 3). Among them is a cDNA (clone PP_LEa0004K19f) encoding a 205 amino acid sequence which shows the signature region of the family 48 glycosyltransferase. The sequence is highly similar to a putative callose synthase 1 catalytic subunit of O. sativa. It has been shown that when plants are attacked by pathogens they respond quickly by depositing callose in damaged sites, suggesting that callose deposition acts as a defence mechanism (Nishimura et al., 2003). Even though peach homologues have not been characterized, the present results open up the possibility that in woolly fruit the response to damage or pathogen infection by callose deposition might be altered. The cDNA clone (PP_LEa0005N23f) encodes a partial sequence of 187 amino acids that shows high similarity to an A. thaliana putative member of the galactosyltransferase family 31. The encoded protein possesses a galectin/galactose-binding domain; such a domain can be found in a number of glycosyltransferases and potentially mediates substrate recognition and/or protein–protein interaction. In eukaryotes, galactosyltransferase family 31 includes β(1→3)-N-acetylglucosaminyltransferases, β(1→3)-galactosyltransferases, β(1→3)-N-acetylgalactosaminyltransferases, and a large number of galactosyltransferases of unknown donor/acceptor specificity. Plant homologues have not been functionally characterized, but high levels of expression of members of this family have been demonstrated in Arabidopsis secondary xylem (Oh et al., 2003) and in hybrid Aspen secondary cell wall biogenesis (Aspeborg et al., 2005). The clone PP_LEa0008K15f encodes a 323 amino acid peptide that showed 75% identity to a putative CCR of A. thaliana (Lauvergeat et al., 2001). In the lignin branch of the phenylpropanoid pathway, CCR is the first enzyme and is responsible for the conversion of hydroxycinnamic acid CoA esters to their corresponding hydroxycinnamaldehydes. The various branches of the phenylpropanoid pathway, biosynthesis of lignin, hydroxycinnamates, and flavonoids, are closely linked, and thus down-regulation of CCR might cause variations in the flux through the pathway leading to the synthesis of a different pool of hydroxycinnamic acids or aldehydes with a putative effect on flavour or on cell wall-bound hydroxycinnamates (Kroon and Williamson, 1999). Consistently, down-regulation of CCR, in tomato, through an RNA interference (RNAi) strategy, leads to quantitative and qualitative changes in the soluble phenolic content of extracts from fruit and vegetative organs and an increase in the antioxidant capacity of the plant extracts (van der Rest et al., 2006).
An additional cDNA encoding a product with potential roles as a structural cell wall protein was also identified as down-regulated in the woolly peach (Table 2). The protein sequence deduced from this cDNA is highly similar to a group of proteins of unknown function described as cotton fibre expressed proteins (from Gossypium hirsutum, accession no. AAC33277). The P. persica protein contains a stretch of hydrophobic residues near the N-terminus, which is similar to a signal sequence commonly seen in secreted proteins. This is supported by the finding of a putative cleavage site that follows the signal sequence in position 22–23. Sequence analysis indicates that this protein is rich in amino acid residues P, V, and S, which are organized in repeats PXV and PPKSV. Thus, the protein encoded by clone PP_LEa0033B07f has some features generally found in cell wall proteins, such as proline richness, repeated motifs, and the presence of a putative signal peptide sequence (Cassab and Varner, 1988; Showalter, 1993; Cassab, 1998). It has been proposed that the interaction of cell wall proteins with the major carbohydrate components of the wall influences cell wall composition and structure (Farrokhi et al., 2006). In peach fruit, the up-regulation of genes encoding cell wall proteins during ripening has led to the suggestion that these proteins contribute to stabilize the wall during the massive dismantling that takes place at fruit softening (Trainotti et al., 2003).
The importance of glycosylphosphatidylinositol (GPI)-anchored proteins (GAPs) for the synthesis and secretion of cell wall polymers has become evident from the analysis of the pnt1 mutant in which a general reduction in GAPs causes remarkable changes in the cellulose content and aberrant deposition of pectin, xyloglucans, and callose (Gillmor et al., 2005). In the present macroarray experiments, a cDNA clone encoding a putative GAP highly similar to Arabidopsis Cob (Schindelman et al., 2001) that was down-regulated in woolly peach fruit was identified (Tables 2, 3). Sequence analysis of the Arabidopsis genome indicated that Cob belongs to a multigene family consisting of 12 members (Roudier et al., 2002). Molecular and genetic studies of the AtCob gene family demonstrated that some of its members are involved in functions such as cell expansion and polarized deposition of cell wall materials. For example, mutations in CobL4 and in its rice homologue brittle culm1 (bc1) gave a severe cellulose-deficient phenotype, indicating that the Cob gene family plays an essential role in cellulose deposition (Schindelman et al., 2001; Roudier et al., 2002; Li et al., 2003). Although Cob proteins have not been characterized in peach, the functional roles attributed to this protein in other systems and the down-regulation of a putative Pp-cob observed in woolly peach suggest that localized deposition of cell wall components may be required during normal ripening. Further analysis should be performed to determine the roles of Cob proteins in peach ripening.
Genes with functions in endomembrane traffic
An interesting response of peach fruit to the woolliness-induced condition was observed for genes potentially encoding proteins with roles in endomembrane trafficking. The plant endomembrane system plays roles in the biogenesis of the cell wall, plasma membrane, and vacuole, and contributes to the control of development and to responses to biotic and abiotic stresses (Surpin and Raikhel, 2004). The major complex polysaccharides (pectin and hemicellulose) of the plant cell wall are synthesized in the Golgi complex and transported by vesicles to the cell wall (Driouich et al., 1993). Cell wall modification enzymes, such as EndoPG and PME, are synthesized on the rough endoplasmic reticulum (ER) (Ray et al., 1988) and mobilized through the cell endomembrane system to be secreted in the apoplast (Staehelin and Moore, 1995). Therefore, the dynamic remodelling of the cell wall requires regulated membrane trafficking, which enables cells to deliver new components to the wall (Johansen et al., 2006). Moreover, endocytosis also appears to be important for cell wall structure (Gillmor et al., 2005) and it has been proposed that cell wall pectins can be recycled from the cell surface (Dhonukshe et al., 2006). In the present macroarray experiments, 11 genes encoding putative proteins with potential functions in the early and late secretory pathways, as well as in the endocytic pathway, showed significant changes of expression between woolly and juicy samples. The expression of nine of these genes was repressed in woolly fruit when compared with juicy fruit (Table 2), and the expression pattern of most of these genes were confirmed by qPCR (Table 3). Our results suggest that alterations in the abundance of the endomembrane system components would have a potentially important role in the acquisition of the woolly phenotype. Certainly these alterations could modify the flow of polysaccharides and proteins to the cell wall.
Among the molecular chaperones that mediate protein folding in the ER, a putative ROC7 cyclophilin encoded by the PP_LEa0009K22f clone that was down-regulated in the woolly fruit was identified (Tables 2, 3). Sequence analysis of Arabidopsis ROC7 has localized it to the ER (Romano et al., 2004) where it might play a role in assisting the proper folding of transmembrane proteins or soluble proteins before they progress along the secretory pathway. Down-regulation of two cDNAs encoding a homologue to the GTPase-activating protein Sec23 and a member of the VAP33 family of SNARE-like proteins, Vap272, suggested that the anterograde ER–Golgi traffic might be affected in the woolly peach (Tables 2, 3). Sec23, which has been detected in association with ER membranes in Arabidopsis cells (Movafeghi et al., 1999), is a component of the COPII coat that stimulates Sar1 GTPase activity (Barlowe and Schekman, 1993; Barlowe et al., 1994; Matsuoka et al., 2001). Sar1 activity, in turn, seems to be required for COPII coat disassembly before vesicle fusion with the target membrane (Oka and Nakano, 1994) and COPII vesicle fission from the ER membrane (Bielli et al., 2005). Characterization of mammalian and yeast homologues of VAP33 has indicated that they localize mainly to ER membranes and suggested that they have a role in vesicle trafficking (Kagiwada et al., 1998; Skehel et al., 2000; Weir et al., 2001). In plants, two members of the family VAP33 have been identified: VAP27 from Nicotiana plumbaginifolia (Laurent et al., 2000) and VAP272 from cowpea (Carette et al., 2002). The latter was located to the ER membrane, and by homology to its mammalian and yeast counterparts it was suggested that VAP272 plays a role in vesicular transport to or from the ER.
Two cDNAs clones encoding putative components of the retrograde protein trafficking from Golgi to ER, gamma 2-COP and ERD2, showed opposite expression patterns (Tables 2, 3). The first of them, clone PP_LEa0004B09f, which encodes a putative gamma 2-COP, a subunit of the COPI complex, exhibited enhanced expression levels in woolly peaches. Gamma 2-Cop has been implicated in the binding of members of the p24 family of transmembrane proteins (Carney and Bowen, 2004). Even though in plant cells there are no experimental data to ascertain which COPI subunits might be responsible for binding to distinct sorting signals, the binding of p24 family proteins to both COPI and COPII has been demonstrated (Contreras et al., 2004) and the expression of different COPI subunits has been affected in several plant species under diverse experimental conditions (Thibaud-Nissen et al., 2003; Martínez and Chrispeels, 2003; Brinker et al., 2004). The second clone (PP_LEa0001H21f) encodes a predicted protein that is highly homologous to ERD2 from A. thaliana, a receptor that cycles between the ER and Golgi and recognizes C-terminal H/KDEL signals of soluble ER resident proteins for sorting into COPI vesicles. The observation that in yeast, mutation in the KDEL receptor resulted in enlarged and disorganized Golgi complexes and altered transport in the early secretory system suggested that KDEL receptors not only retrieve proteins, but also regulate intracellular trafficking (Semenza et al., 1990). In Arabidopsis, ERD2–GFP (green fluorescent protein) accumulated at Golgi stacks of tobacco cells, as expected for a protein with functions in the Golgi to ER recycling (Boevink et al., 1998), and its expression was highly increased in secretory tissues (Bar-Peled et al., 1995). Further analysis will be needed to assess the concerted or antagonist role of these proteins during the ripening process.
The pathway from the trans-Golgi network (TGN) to the plasma membrane transports cell wall polysaccharides and both structural and enzymatic proteins to the cell surface. In this work, two genes were identified as repressed in the woolly peaches that encode putative R-SNARE and Rab11 proteins, which might control membrane fusion of secretory vesicles with the plasma membrane (Tables 2, 3). Clone PP_LEa0014G24f encodes a putative vesicle-associated membrane protein 722 (VAMP722) that possesses 82% identity with the Arabidopsis VAMP722 (accession no. P47192). Two groups of VAMP7 proteins have been described in Arabidopsis: AtVAMP71 (AtVAMP711–AtVAMP714) and AtVAMP72 (AtVAMP721–AtVAM727). The VAMP72 group appears to be specific to the green plant lineage and it was proposed to function as the R-SNARE component for secretion (Sanderfoot, 2007). Consistently, several VAMP72s, including VAMP722, have been localized to the plasma membrane (Marmagne et al., 2004; Uemura et al., 2005). The clone PP_LEa0020B02f encodes a putative P. persica Rab11a GTPase isoform, which has been involved in ripening in other species. A Rab11a cDNA was isolated from mango (Mangifera indica L.) as a sequence differentially expressed between ripe and unripe fruit (Zainal et al., 1996). Moreover, in tomato [Solanum lycopersicum (formerly named Lycopersicon esculentum)], the expression of Rab11 was induced during ripening, and antisense inhibition of Rab11 resulted in reduced levels of the two main cell wall-modifying enzymes, PG and PME, suggesting a possible role in trafficking of cell wall-modifying enzymes (Lu et al., 2001). Interestingly, the expression of LeRab11a was repressed in the Nr tomato mutant, which is unable to perceive ethylene because of a mutation in an ethylene receptor (Wilkinson et al., 1995), suggesting that LeRab11a is regulated in an ethylene-dependent manner in fruit. Under the assumption that woolliness can be the result of an abnormal ripening, these data suggest that a down-regulation of PpRab11a in woolly peach fruit would affect the normal secretion of cell wall components during ripening, and open up the possibility that PpRab11a repression might be the result of the low ethylene evolution that takes place in woolly peaches.
Four genes encoding putative proteins with roles in endocytosis were also identified as differentially expressed between juicy and woolly peaches (Tables 2, 3). Among them is a product (encoded by PP_LEa0009I13f) with a dynamin GTPase domain, which is highly homologous to ADL1A (Arabidopsis dynamin-like protein 1A) and Glycine max phragmoplastin. These two dynamin-related proteins lack the pleckstrin and proline-rich domains characteristic of mammalian dynamin I and are expected to perform plant-specific functions (Praefcke and McMahon, 2004). It has been proposed that ADL1 mediates cell plate assembly, cell wall formation, and plasma membrane recycling (Kang et al., 2003a, b). A probable role for ADL1 in endocytosis has been suggested by its ability to co-purify with auxin efflux protein complexes (Murphy et al., 2002). Prunus persica clone PP_LEa0013L18f encodes a putative ARA7/RabF2b, a member of the Ypt51p/Rab5 family of small GTP-binding proteins. In mammals, Rab5 GTPases regulate the early steps of endocytosis (Bucci et al., 1992). Similarly, yeast Ypt51p family members regulate membrane trafficking through pre-vacuolar compartments (Horazdovsky et al., 1994; Singer-Krüger et al., 1995). Arabidopsis ARA7, which is a close homologue of mammalian Rab5, has been co-localized in early endosomal compartments with the styryl dye FM 4-64, a marker for the endocytic pathway (Ueda et al., 2001). Moreover, constitutive active mutants of Ara7 form aggregates of deformed endosomes, suggesting that ARA7, like its animal counterpart, acts as a regulator of endosome membrane fusion (Ueda et al., 2001). The clone PP_LEa0017D15f encodes a product with homology to β-adaptin, a component of the clathrin-recruiting AP-2 complex. Plant β-adaptins have been identified in zucchini (Holstein et al., 1994) and tentatively localized at the plasma membrane (Drucker et al., 1995). β-Adaptins were also identified as components of plasma membrane complexes containing Arabidopsis auxin efflux proteins (Murphy et al., 2002). Even though it has been assumed that plant β-adaptins may function analogously to mammalian β-adaptins, their roles in clathrin binding to AP-2 complexes have not been experimentally demonstrated. The last member of the four genes potentially involved in endocytosis was represented by clone PP_LEa0030L15f, which encodes a putative secretory carrier membrane protein (SCAMP); this was also identified as down-regulated in the present screening. SCAMPs are conserved integral membrane proteins that have been implicated in regulatory vesicle trafficking (Hubbard et al., 2000) and localized in both the TGN and recycling endosomes (Castle and Castle, 2005). The main function ascribed to SCAMPs has been to facilitate clathrin-mediated endocytosis (Fernandez-Chacon and Südhof, 2000). Plant SCAMPs have not been functionally characterized; however, SCAMP1 has been identified in tobacco BY-2 cells as localized to both plasma membrane and early endosomes (Lam et al., 2007), whereas putative SCAMPs have been identified in Arabidopsis plasma membrane (Alexandersson et al., 2004; Marmagne et al., 2004). The importance of endocytic events in cell wall remodelling has become apparent from studies showing that cell wall pectins can be recycled from the surface and inserted into the growing cell plate during cytokinesis (Dhonukshe et al., 2006). Moreover, cells undergoing either division or rapid growth internalize large amounts of pectins and GPI-anchored arabinogalactan proteins, whereas pectins that are cross-linked with boron and calcium are internalized and apparently recycled via early endosomes (Baluska et al., 2002, 2005). These results indicate that cell wall substrates can be supplied by endocytosis when rapid remodelling of walls is needed; however, the role of endocytic delivery of cell wall materials in other processes such as fruit ripening or woolliness needs to be examined.
The transcriptional changes listed in this study suggest that molecular changes that take place during fruit woolliness include not only cell wall-degrading enzymes but also a number of genes associated with membrane trafficking within the secretory and endocytic pathways. In this regard, studies on the Cnr mutant of tomato which shows a reduced intracellular adhesion and a woolly texture have revealed that deposition and/or secretion of cell wall arabinan is disrupted (Orfila et al., 2001), connecting a defect in the secretory mechanisms with an altered composition of cell wall polysaccharides.
The data reported here provide information for a better understanding of gene expression changes during development of peach woolliness and they can be extrapolated to other fruit of the Rosaceae family. The genes identified in this study and verified by qPCR are potential targets for future research on the molecular basis of peach woolliness and may yield new traits for development of plants with higher agronomic value.
This work was supported by FDI G02P1001 (Chilean Genome Initiative), ASOEX (Asociación de Exportadores de Chile A.G.), FDF (Fundación para el Desarrollo Frutícola), and Fundación Chile; Fondecyt 1050235 to VC; MGA hold a CONICYT fellowship (AT- 24050114) and an Abraham Stekel fellowship from Nestle Chile-INTA.
References
Author notes
Present address: Institute of Agricultural Research (INIA-La Platina), Santiago, Chile.



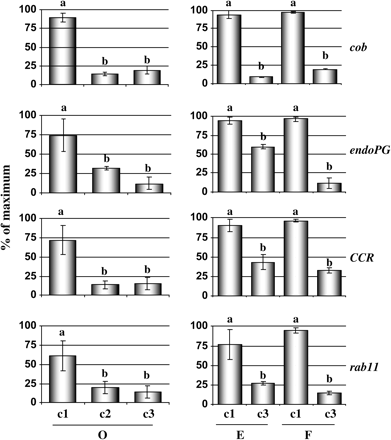

Comments