-
PDF
- Split View
-
Views
-
Cite
Cite
Masanobu Nakamura, Tatsuro Satoh, Shin-Ichiro Tanaka, Nobuyoshi Mochizuki, Takao Yokota, Akira Nagatani, Activation of the cytochrome P450 gene, CYP72C1, reduces the levels of active brassinosteroids in vivo, Journal of Experimental Botany, Volume 56, Issue 413, March 2005, Pages 833–840, https://doi.org/10.1093/jxb/eri073
Close - Share Icon Share
Abstract
A pool of Arabidopsis lines transformed with the activation vector was screened for short hypocotyl mutants under dim far-red light, and three mutant lines designated chibi1–3 (chi) were isolated. Among the chi mutants, chi2 was dominant. The chi2 seedlings were short, regardless of the light conditions. The chi2 mature plants exhibited phenotypic features such as dwarfism, reduced male fertility and dark green, rounded epinastic leaves, which are characteristics of brassinosteroid-deficient mutants such as det2, cpd, and dwf4. Furthermore, the hypocotyl phenotype was restored by the addition of brassinolide to the culture medium, suggesting that brassinosteroids had been affected in this mutant. The molecular analysis of the chi2 mutant revealed that the CYP72C1 gene was overexpressed by the enhancing activity of the inserted DNA. Wild-type plants that were transformed with a vector containing a chimeric gene between the 35S promoter and the CYP72C1 genomic DNA exhibited a similar phenotype. Consistent with the morphological and physiological phenotype, the levels of active brassinosteroids were reduced in the chi2 mutant. Hence, CYP72C1, together with BAS1/CYP72B1, is speculated to regulate active brassinosteroid levels in plants. Expression analysis suggested that wild-type CYP72C1 transcript levels increased after exposure to white light, although the physiological significance of such a response remains obscure.
Introduction
Plant growth and development are regulated by interactions between environmental and endogenous developmental signals, and light is one of the most important environmental signals. Activation of photoreceptors in plants leads to various physiological as well as developmental responses such as germination, de-etiolation, shade avoidance, and flowering (Fankhauser and Chory, 1997). Higher plants have evolved a complex photoreceptive system involving distinct families of photoreceptors to perceive light signals (Kendrick and Kronenberg, 1994).
In the responses of plants to light, phytohormones act as endogenous signals. The involvement of hormones such as brassinosteroid (BR), auxin, and ethylene in seedling de-etiolation triggered by light has been proposed (O'Brien et al., 1985; Kraepiel and Miginiac, 1997), and this view is supported by recent genetic analysis. Some of the mutants that exhibit the developmental characteristics of light-grown plants in complete darkness have turned out to be BR-deficient (Szekeres et al., 1996; Altmann, 1999; Chory and Li, 1997; Clouse, 1997; Szekeres and Koncz, 1998; Yokota, 1997). This suggests that BR acts as a negative regulator of photomorphogenesis (Nemhauser and Chory, 2004).
BRs are plant-specific steroids, which regulate various aspects of growth including stem elongation, leaf unrolling, and stress responses (Bishop and Yokota, 2001). The BR family comprises more than 30 different steroids. Among these, brassinolide, the most biologically active BR, is distributed widely in the plant kingdom together with its precursors. Detailed analysis with cultured Catharanthus roseus cells revealed that brassinolide is synthesized from castasterone, generated from campestanol, via either the early or late C6-oxidation pathways (Fujioka and Yokota, 2003). These pathways also contribute to the synthesis of active BRs in Arabidopsis (Fujioka and Yokota, 2003). Several dwarf mutants of Arabidopsis have lower levels of BRs. Cloning of the responsible genes for these mutants has resulted in the identification of many BR biosynthetic enzymes including the cytochrome P450s (CYPs) (Altmann, 1998).
Inactivation of steroid hormones is important for proper development in animals (for review, see Gade et al., 1997; Omdahl et al., 2002; Michael et al., 2003). In insects, rapid inactivation of a steroid hormone, ecdysteroid, leads to moulting. CYP enzymes play a role in steroid inactivation; for example, vitamin D, which is widely distributed in animals, is inactivated by a CYP. In plants, while various BR metabolites have been identified (Fujioka and Yokota, 2003), little is known about the enzymes involved in BR metabolism. BAS1/CYP72B1 is the only enzyme that is known to catabolize active BRs in plants (Neff et al., 1999).
The CYP family in Arabidopsis consists of 246 members (Yamanaka et al., 2003). They oxidize diverse substrates such as membrane sterols, lignin, and cutin as well as hormones. Some members are involved in the biosynthesis of secondary compounds, such as flavonoids, isoprenoids, and alkaloids, but the biological functions of other members remain unclear at present. The CYP72 family consists of three subtypes, CYP72A, B, and C (Werck-Reichhart et al., 2002). Although CYP72B and CYP72C genes exist as single genes, there are eight CYP72A-type genes (CYP72A7–15) in the Arabidopsis genome. In Catharanthus, the involvement of CYP72A1 in the biosynthetic pathway for the terpene indole alkaloids has been reported (Irmler et al., 2000).
The activation-tagging strategy (Hayashi et al., 1992) has been successfully applied to the study of photomorphogenesis. Plants have been transformed with a vector containing multiple copies of the 35S enhancer, with the expectation that genes in the vicinity of the transferred-DNA (T-DNA) insertion sites will be activated (Walden et al., 1994). Using this strategy, mutants that exhibit partial photomorphogenesis in darkness have been isolated. Analysis of one such mutant, bas1, revealed that overexpression of a member of the CYP72 gene family, BAS1/CYP72B1, resulted in a BR deficiency (Neff et al., 1999). The authors have proposed that BAS1/CYP72B1 hydroxylates active BRs such as brassinolide and castasterone (Turk et al., 2003). The recent analysis of the loss-of-function bas1/cyp72b1 mutant suggested that the BAS1/CYP72B1 protein acts as a positive regulator of photomorphogenesis under the control of phytochrome A, which is the photoreceptor regulating hypocotyl growth under continuous far-red light (Turk et al., 2003).
In the present study, an activation-tagged Arabidopsis pool was screened for short hypocotyl mutants under dim far-red light and a dominant dwarf mutant named chibi2 (chi2) was isolated. Molecular analysis of this mutant revealed that the CYP72C1 gene, a member of the CYP family, was over-expressed. Quantification of BRs indicated that the levels of active BRs were severely reduced in this mutant. Hence, in this paper the possible role of CYP72C1 in plant BR metabolism is discussed.
Materials and methods
Plant materials and growth conditions
Establishment of the activation-tagging lines has been described elsewhere (Nakamura et al., 2000). In brief, Arabidopsis thaliana (ecotype Ler) plants were transformed with the activation vector pPCVICEn4HPT containing four copies of the viral 35S enhancer (Hayashi et al., 1992).
For seedling experiments and mutant screening, seeds were sown on 0.6% agar plates containing the Murashige and Skoog salt mixture (Wako, Tokyo) without sucrose. Plants were further grown on vermiculite soil under white fluorescent tubes to observe mature plants and to harvest seeds. To examine the effects of phytohormones on the hypocotyl elongation in darkness, seedlings were grown in the presence of brassinolide at 10−10−10−7 M and GA3 at 10−9−10−6 M.
Light treatments
Far-red light was obtained from colour fluorescent tubes (FL20S-FR74, Toshiba, Tokyo) filtered through a 3 mm methacrylate plate (Delaglass A-900, Asahi Chemical Industry). Blue light was obtained from blue fluorescent tubes (FL20S/B-F, National) wrapped with blue film (No. 72, Tokyo Butai Showmei, Tokyo). The red light source has been described elsewhere (Yamaguchi et al., 1999). Monochromatic lights were attenuated with a grey plastic plate if necessary (Takiron plates S-802 and S-909, Takiron, Tokyo). Light intensities were measured with a DataLogger (Li-Cor, Lincoln, Nebraska).
Genetic and molecular biological analysis
The thermal asymmetric interlaced-polymerase chain reaction (TAIL-PCR) was employed to clone the Arabidopsis (Ler ecotype) genome region flanking the inserted T-DNA sequence (Liu and Whittier, 1995). An approximately 3.5 kb Spe I-Xba I fragment of the Arabidopsis genomic DNA corresponding to the full length of the CYP72C1 coding region was ligated to XbaI site of pBI-Hm/35S-nosT (Yamaguchi et al., 1999) to obtain a vector for CYP72C1 overexpression. The vector was transformed into the wild-type Arabidopsis (ecotype Col-0) by the in planta transformation method with the following minor modification (Bechtold et al., 1993; Yamaguchi et al., 1999): the duration and the air pressure for the vacuum infiltration were varied from 5 min to 15 min and from 100 mm Hg to 400 mm Hg, respectively.
RNA gel blot analysis and RT-PCR analysis
For RNA gel blot hybridization, a CYP72C1-specific probe corresponding to the second and third exons was prepared using [α-32P] dCTP (800 Ci mmol−1, Amersham-Japan, Tokyo) and the BcaBest Labeling Kit (Takara, Japan). Total RNA isolation was performed using the RNeasy Plant RNA Isolation Kit (Qiagen, Hilden, Germany). For the reverse transcription-polymerase chain reaction (RT-PCR) analysis, RNA was extracted from 6-d-old seedlings grown under continuous white light or in complete darkness as previously described (Tanaka et al., 2002). A 1 μg sample of isolated total RNA was treated with DNase (Promega, CA, USA) and subjected to the reverse transcription reaction (Invitrogen, CA, USA). The resulted cDNA was diluted using the polyubiquitin 6 (UBQ6) gene as an internal control.
Determination of brassinosteroid levels
Levels of BRs in the chi2 mutant and the wild-type plants were determined as previously described (Mori et al., 2002; Nomura et al., 2001). Methanol extracts of 24-d-old plants (average fresh weights, 56 g (chi2) and 191 g (WT)) were spiked with 0.1 μg each of 2H6-labelled BRs (1 μg for 2H6-labelled 6-deoxocastasterone) supplied by Dr S Takatsuto, prior to reduction to an aqueous residue. The ethyl acetate-soluble neutral fraction was obtained from the residue and partitioned between 80% methanol and hexane. The former portion was further purified successively on a silica gel column (Wakogel C-300, 7 g; Wako Pure Chemicals, Osaka, Japan), a Sephadex LH-20 column (bed volume, 500 ml; Pharmacia Fine Chemicals, Uppsala, Sweden), and a charcoal column (chromatography grade, 5 g; Wako Pure Chemicals, Japan). BRs were then chromatographed by reversed-phase HPLC on a SenshuPak ODS-3251-D column (8× 250 mm; Senshu Science, Tokyo, Japan). Separated BRs were quantified by gas chromatography-selected ion monitoring (GC-SIM) after converting to either monomethaneboronates, bismethaneboronates or monomethaneboronate trimethylsilyl ethers.
For sterol determination, a random aliquot (50 mg of fresh weight equivalent) of the above-described ethyl acetate-soluble fraction was sampled and spiked with 1 μg of [2H6]-campestanol as an internal standard. The hydrolysate obtained by saponification with methanolic 1 N KOH was diluted with water and extracted with hexane. The hexane extract was evaporated, redissolved in chloroform and passed through a short silica gel column (Wakogel C-300). The eluate was trimethylsilylated at room temperature and subjected to GC-SIM quantitation as described by Nomura et al. (2001).
Results
Isolation of the chi2 mutant
To understand the mechanism of hypocotyl de-etiolation better, Arabidopsis was screened for mutants in which hypocotyl elongation was severely inhibited. Seedlings of 2500 activation-tagged Arabidopsis lines (ecotype Ler) were grown under dim far-red light of 0.35 W m−2 for 6 d on agar plates. The lines that exhibited short hypocotyl phenotype were further analysed, and three independent mutant lines designated as chibi1 to chibi3 (chi1–3) were identified. In the segregating T2 population of the chi mutants, short seedlings were recognized in a 1:3 ratio (mutant/wild-type) in chi1 and chi3, whereas a 3:1 segregation was observed for the chi2 mutant, indicating the dominant nature of the chi2 mutation (Fig. 1A). This was further confirmed by crossing the homozygous chi2 mutant with the wild-type plant. 142 short plants in 186 F2 siblings were observed.
Photographs of the chi2 plants. (A) Segregating the T2 siblings of the chi2 mutant. Seedlings were grown under dim far-red light (0.35 W m−2) for 6 d. Scale bar=5 mm. (B) The wild-type (Ler) plant grown for 27 d. Scale bar=1 cm. (C) A mature chi2 plant grown for 27 d. Scale bar= 1 cm. (D) The Arabidopsis plant transformed with the 35S∷CYP72C1 construct. The plants were grown for 30 d. Scale bar=1 cm. (E) The Arabidopsis plant transformed with the control vector. The plants were grown for 30 d. Scale bar=1 cm. (F–I) The wild-type (F, H) and the chi2 (G, I) seedlings grown for 6 d in darkness. Seedlings were grown in the absence (F, G) or presence (H, I) of 10−7 M brassinolide.
At the mature stage, the chi2 mutant resembled BR-deficient mutants such as cbb1/dim/dwf1–6 (Choe et al., 1998), cbb2/bri1 (Li and Chory, 1997), cbb3/cpd (Choe et al., 1998), det2 (Li et al., 1996), and bas1 (Neff et al., 1999). Similar to these mutants, chi2 was severely dwarfed compared with the wild-type plants (Fig. 1B, C). In addition, reduced male fertility and dark green, rounded and epinastic leaves were observed. These observations indicated reduced levels of active BRs in this mutant.
Physiological characterization of the chi2 mutant
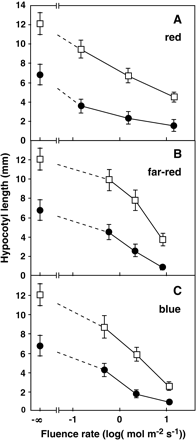
The homozygous chi2 mutant was further characterized under different light conditions. Seedlings were grown on agar plates under constant light conditions for 6 d. In darkness, chi2 (7.0±1.1 mm) was significantly shorter than the wild-type plants (12.2±1.2 mm) (Fig. 1F, G). The apical hook of the chi2 seedling opened in darkness. The hypocotyl phenotype was then examined under monochromatic red, blue, and far-red lights (Fig. 2). As shown in the figures, the chi2 mutant retained the ability to respond to light, while both the responsiveness and sensitivity of the response appeared to have increased. These phenotypic features have been reported in other brassinosteroid-deficient mutants (Chory et al., 1989, 1991; Szekeres et al., 1996).
Hypocotyl responses of the chi2 mutant to monochromatic lights. Hypocotyl lengths of the chi2 (filled circles) and the wild-type (open squares) seedlings were examined. Seedlings were grown under different fluence rates of continuous red (A), far-red (B) or blue (C) lights for 6 d. Averages of three independent experiments are shown. Error bars indicate SD.
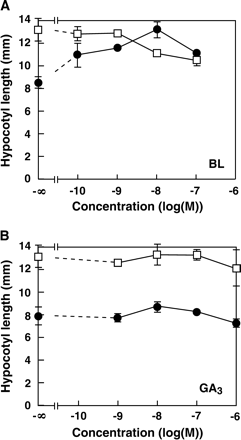
To test whether the addition of hormones would rescue the chi2 mutation phenotype, chi2 seedlings were grown in the presence of exogenous brassinolide or giberellin (Fig. 3). Addition of over 10−8 M brassinolide to the medium had an inhibitory effect on the wild-type hypocotyl elongation in darkness (Fig. 3A). At lower concentrations, brassinolide did not affect the hypocotyl elongation. By contrast, brassinolide at 10−10−10−8 M promoted the hypocotyl growth in the chi2 mutant, indicating that the short hypocotyl phenotype of the chi2 seedling was rescued by the addition of brassinolide. GA3 treatment was also examined, but it affected growth only slightly in both the wild-type and chi2 seedlings (Fig. 3B).
Effects of brassinolide and GA3 on hypocotyl elongation in darkness. Hypocotyl lengths of the chi2 (filled circles) and the wild-type (open squares) seedlings were examined. Seedlings were grown in the presence of different concentrations of brassinolide (upper) or GA3 (lower) for 6 d. Averages of two independent experiments are shown. Error bars indicate SD.
Genetic and molecular biological analysis of the chi2 mutant
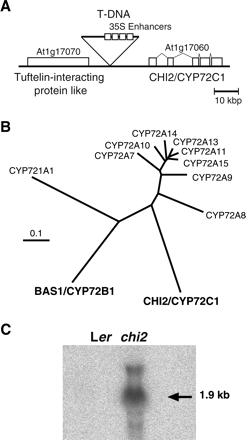
The chi2 mutation was roughly mapped to 20 cM on chromosome 1 by simple sequence length polymorphism (SSLP) (Bell and Ecker, 1994) and cleaved amplified polymorphic sequence (CAPS) (Konieczny and Ausbel, 1993) analyses. An approximately 3 kb portion of the genome DNA adjacent to the left border of the T-DNA was cloned by TAIL-PCR and sequenced. The results indicated that the T-DNA was inserted between two hypothetical open reading frames, At1g17060 and At1g17070 (Fig. 4A). Interestingly, At1g17060 exhibits homology to the CYPs. According to the CYP nomenclature committee, this open reading frame was designated as CYP72C1. Phylogenic analysis revealed that both CYP72C1 and BAS1/CYP72B1 belong to a relatively small clade in the CYP family (Fig. 4B).
Molecular biological analysis of the chi2 mutant. (A) The T-DNA insertion point on chromosome 1 in the chi2 mutant. The activation vector contains four copies of the 35S enhancers. (B) A gene tree of a clade of the Arabidopsis CYP genes including the CHI2/CYP72C1 and the BAS1/CYP72B1 genes. All members from the Arabidopsis CYP72 subfamily and CYP721A1 protein sequences are included in the tree. Amino acid sequences registered in MIPS database were analysed with the Clustal W program. (C) RNA gel blot analysis of the CYP72C1 gene expression in the chi2 plants. RNA was extracted from the aerial parts of the wild-type and the chi2 plants grown for 30 d. The blot was hybridized with a probe corresponding to the second and third exons of the CHI2/CYP72C1 gene.
Northern blot analysis was performed to confirm that CYP72C1 was overexpressed in the chi2 mutant (Fig. 4C). RNA was isolated from the aerial part of plants grown for 30 d in white light. The gel blot was hybridized with a CYP72C1 specific probe corresponding to the second and the third exons of the gene. In these results, an intense CYP72C1 mRNA signal was detected in the chi2, but not in the wild-type RNA preparation. To confirm that CYP72C1 was indeed the causal gene for the chi2 mutation, the full length CYP72C1 cDNA was cloned from the chi2 mutant plants and transformed into the wild-type Arabidopsis (ecotype Col-0). The obtained transgenic plants exhibited phenotype indistinguishable from that of the original chi2 mutation, although the leaf epinasty was less clear (Fig. 1D). By contrast, transgenic plants transformed with the control vector never exhibited dwarf phenotype (data not shown). On the basis of these findings, it was concluded that CYP72C1 is the causal gene for the chi2 mutation.
Brassinosteroid levels in the chi2 mutant
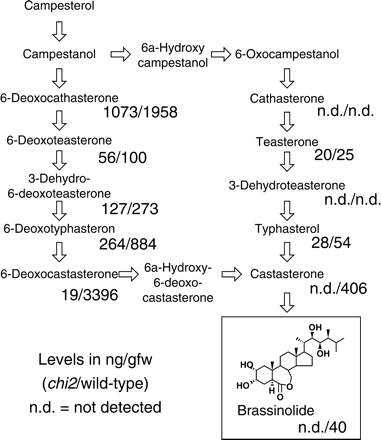
The levels of sterols and BRs in the chi2 and wild-type plants were determined (Fig. 5). The contents (ng g−1 FW) of sterols in chi2, including campesterol (77), campestanol (2), isofucosterol (19), sitosterol (357), stigmasterol (4.5), sitostanol (6), and cholesterol (31) were not significantly varied from those of wild-type plants. However, the levels of all the BRs detected were reduced to some extent in the chi2 mutant (Fig. 5). It is remarkable that castasterone and brassinolide were not present in the chi2 mutant. Brassinolide is the biologically active BR, and its direct precursor castasterone is also considered to be active (Fujioka and Yokota, 2003). Hence, the levels of active BRs were severely reduced by the overexpression of CHI2/CYP72C1 in the chi2 mutant. Furthermore, it is also remarkable that the level of 6-deoxocastasterone, the precursor of castasterone, was reduced to only 0.6% of that in wild-type plants.
BR content in the chi2 and the wild-type plants. A proposed biosynthetic pathway for brassinolide (Fujioka and Yokota, 2003) is shown. The levels in ng g−1 FW are indicated below the names of BRs (chi2 mutant/wild-type). nd=not detected.
The CHI2/CYP72C1 gene expression in the wild-type seedlings
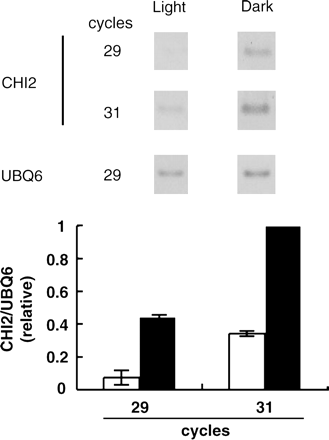
To investigate the actual function of CHI2/CYP72C1, the level of its mRNA was examined. Total RNA was isolated from seedlings grown under continuous white light or in darkness, and this was subjected to RT-PCR analysis (Fig. 6). DNA staining of the samples at 29 and 31 cycles is shown. In both cases, the sample from dark-grown seedlings exhibited a higher signal than that from the light-grown seedlings. Hence, the expression of CHI2/CYP72C1 was down-regulated by light.
RT-PCR analysis of the CYP72C1 gene expression in the wild-type plants. The CHI2/CYP72C1 gene expression is shown for light-grown (left) and dark-grown (right) seedlings. The loading control (UBQ6) is shown in the bottom row. The result of the quantification of the data is shown in the lower panel. Averages of three independent experiments are shown. Error bars indicate SD.
Discussion
The present work has defined a novel function for CHI2/CYP72C1. Biologically active BRs such as castasterone and brassinolide vanished when CHI2/CYP72C1 was overexpressed in the chi2 mutant, suggesting that CHI2/CYP72C1 catabolizes the active BRs. Recently, BAS1/CYP72B1 has been demonstrated to reduce the levels of castasterone and brassinolide by conversion to their 26-hydroxy derivatives (Turk et al., 2003). CHI2/CYP72C1 and BAS1/CYP72B1 share 47% identity and 66% homology. The putative haem-binding domain surrounding the ERR-motif and a transmembrane domain are conserved in both the CHI2/CYP72C1 and BAS1/CYP72B1 sequences. Thus, it is likely that these genes have a common function.
CHI2/CYP72C1 and BAS1/CYP72B1 may catabolize 6-deoxocastasterone in addition to castasterone and brassinolide, because the level of 6-deoxocastasterone is severely reduced in both the chi2 and bas1-D mutants (Neff et al., 1999). It was recently discovered that the chi2 mutant metabolizes radiolabelled 6-deoxocastasterone more rapidly than do wild-type plants (T Yokota and T Sato, unpublished results). Taken together, it seems that CHI2/CYP72C1 and BAS1/CYP72B1 have a broader substrate specificity than expected. It should be noted here that the substrate preferences of these two CYPs may not be identical. The levels of all brassinosteorids including 6-deoxoteasterone were reduced in the chi2 mutant (Fig. 5), whereas the level of 6-deoxoteasterone was found to be increased in bas1-D mutants (Neff et al., 1999).
There are numerous CYPs in Arabidopsis (Yamanaka et al., 2003), and they play crucial roles in the biosynthesis of a variety of endogenous lipophilic compounds such as fatty acids, sterols, phenylpropanoids, terpenoids, phytoalexins, and gibberellins (Werck-Reichhart et al., 2002). In addition, oxidative detoxification of a number of herbicides in plant tissues is also achieved by a CYP-dependent mono-oxygenase system (Mizutani et al., 1998). A phylogenic analysis indicated that CHI2/CYP72C1 and BAS1/CYP72B1 proteins are present in a relatively small clade of the Arabidopsis CYP family (Fig. 4B). Hence, other members of the clade may have capabilities similar to those of CHI2/CYP72C1 and BAS1/CYP72B1.
It is interesting that the CHI2/CYP72C1 gene was down-regulated by light (Fig. 6) and, as discussed above, CHI2/CYP72C1 serves to reduce the levels of active BRs. BAS1/CYP72B1 gene expression is also down-regulated by light (Turk et al., 2003); hence, these genes may be acting together to inactivate BRs in darkness. This fits well with the observation that BR levels are higher in light-grown versus dark-grown seedlings in pea (Symons et al., 2002) and rice (Fujioka and Yokota, 2003). However, these and other findings cast doubt on whether BRs act as negative regulators of photomorphogenesis (Symons and Reid, 2003).
The phenotype of the bas1/cyp72b1 null mutant has been examined recently (Turk et al., 2003). Interestingly, no long hypocotyl phenotype was observed for this mutant in the absence of light, despite the fact that BR levels should be increased in the dark. It is noteworthy that the bas1/cyp72b1 mutant exhibits the most prominent long hypocotyl phenotype under far-red light. Expression levels of the BAS1/CYP72B1 gene are as high under far-red light exposure as in darkness. Hence, BAS1/CYP72B1 may be involved in the inhibition of hypocotyl elongation after far-red light exposure (Turk et al., 2003). Considering the similarity between the CHI2/CYP72C1 and BAS1/CYP72B1 proteins, CHI2/CYP72C1 may also play a similar role in vivo. Further analysis of the loss-of-function chi2/cyp72c1 mutant is needed.
The CHI2/CYP72C1 gene may have additional functions unrelated to hypocotyl growth. BRs are known to be required for vasculature formation (Yamamoto et al., 1997), seed germination (Ullah et al., 1997; Chen et al., 2004), and root development (Mussig et al., 2003). Interestingly, data from AtGenExpress indicate strong expression of the CHI2/CYP72C1 gene in roots and siliques, with higher transcript levels in seeds (https://www.genevestigator.ethz.ch/). Hence the CHI2/CYP72C1 gene may be regulating BR levels in these organs.
Genes such as CHI2/CYP72C1 and BAS1/CYP72B1 should be useful in the genetic manipulation of BR levels in plants. In combination with specific promoters, it should be possible to reduce the levels of active BRs in specific organs/tissues. In this way, it might be possible to manipulate BR levels in specific parts of the plant to improve crop plants (Bishop, 2003). This technique would be applicable not only to dicots but also to monocots because active BRs are common to both groups. Hence, many crop plants can be good targets for such manipulation. Furthermore, genome projects on various crop species have revealed many cytochrome P450 genes, and functional studies of these CYPs are anticipated. It has recently been found that the garden pea has a CYP72B homologue, but none for CYP72C (T Yokota and T Nomura, unpublished results). No CYP72B or CYP72C homologues have been identified in other plant species to date. However, CYP72A homologues exist in rice, maize, wheat, tomato, and Solanum spp. (see Directory of P450-containing Systems, http://www.icgeb.org/∼p450srv/biblioD.html). In particular, there are as many as nine CYP72A family genes (CYP72A17 to CYP72A25) in the rice genome, which may also be involved in BR catabolism.
Abbreviations: BR, brassinosteroid; CYP, cytochrome P450; CHI2, chibi2; GC-SIM, gas chromatography and selected ion monitoring; RT-PCR, reverse transcription-polymerase chain reaction; T-DNA, transferred-DNA; UBQ6, polyubiquitin 6.
This work was supported, in part, by Grants-in-Aid for Scientific Research (B) (nos 13440239, 15370020), Grant-in-Aid for Scientific Research (grant no.15004753), a Grant-in-Aid for Scientific Research on Priority Areas (2) ‘Studies on Photoperception and Signal Transduction Pathways of the Blue Light Receptor, PHOT, in plants’ (no. 13139201) and a Grant-in-Aid for 21st Century COE Research, Kyoto University (A14), from the Ministry of Education, Sports, Culture, Science, and Technology of Japan. We thank NASC and AIMS for providing the seeds for the promoter/enhancer trap lines. TY was supported by a grant-in-aid for scientific research from the Japan Society for the Promotion of Science (grant no. 1146007) and was also funded by Human Frontier Research Program (grant no. 2000-162).
References
Altmann T.
Altmann T.
Bechtold N, Ellis J, Pelletier G.
Bell CJ, Ecker JR.
Bishop GJ, Yokota T.
Chen JG, Pandey S, Huang J, Alonso JM, Ecker JR, Assmann SM, Jones AM.
Choe S, Dilkes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA.
Chory J, Li J.
Chory J, Peto C, Feinbaum R, Pratt L, Ausbel F.
Chory J, Nagpal P, Peto C.
Clouse SD.
Fankhauser C, Chory J.
Fujioka S, Yokota T.
Gade G, Hoffmann KH, Spring JH.
Hayashi H, Czaja I, Lubenow H, Schell J, Walden R.
Irmler S, Schroder G, St-Pierre B, Crouch NP, Hotze M, Schmidt J, Strack D, Matern U, Schroder J.
Kendrick RE, Kronenberg GHM.
Kraepiel Y, Miginiac E.
Konieczny A, Ausubel FM.
Li J, Nagpal P, Vitart V, McMorris TC, Chory J.
Li J, Chory J.
Liu YG, Whittier RF.
Michael AE, Thurston LM, Rae MT.
Mizutani M, Ward E, Ohta D.
Mori M, Nomura T, Ooka H, et al.
Mussig C, Shin GH, Altmann T.
Nakamura M, Mochizuki N, Nagatani A.
Neff MM, Nguyen SM, Malancharuvil EJ, et al.
Nemhauser JL, Chory J.
Nomura T, Sato T, Bishop GJ, Kamiya Y, Takatsuto S, Yokota T.
O'Brien T, Beall FD, Smith H.
Omdahl JL, Morris HA, May BK.
Symons GM, Reid JB.
Symons GM, Schultz L, Huub L, Kerckhoffs J, Davies NW, Gregory D, Reid JB.
Szekers M, Koncz C.
Szekers M, Németh K, Koncz-Kálman Z, Mathur J, Kauschmann A, Altmann T, Rédei GP, Nagy F, Schell J, Koncz C.
Tanaka S, Mochizuki N, Nagatani A.
Turk EM, Fujioka S, Seto H, Shimada Y, Takatsuto S, Yoshida S, Denzel MA, Torres QI, Neff MM.
Ullah H, Chen JG, Wang S, Jones AM.
Walden R, Fritze K, Hayashi H, Miklashevichs E, Harling H, Schell J.
Werck-Reichhart D, Bak S, Paquette S.
Yamanaka S, Suzuki E, Tanaka M, Takeda Y, Watanabe JA, Watanabe KA.
Yamaguchi R, Nakamura M, Mochizuki N, Nagatani A.
Yamamoto R, Demura T, Fukuda H.





Comments