-
PDF
- Split View
-
Views
-
Cite
Cite
Brian G. Forde, Is it good noise? The role of developmental instability in the shaping of a root system, Journal of Experimental Botany, Volume 60, Issue 14, October 2009, Pages 3989–4002, https://doi.org/10.1093/jxb/erp265
Close - Share Icon Share
Abstract
Root architecture plays a major part in determining a root system's ability to function effectively and efficiently in its essential roles of anchorage and the capture of soil resources. The characteristics of root development that are conventionally considered to be the main determinants of root architecture are the rate, angle, and duration of root growth and the pattern of root branching. In this review, the case is made that there is an additional trait that has been largely ignored but which has a significant influence on root architecture, namely the degree to which stochasticity (or ‘developmental instability’) affects the developmental process. Although the intrinsic variability in the development and growth of lateral roots has been recognized for some time, in almost every study of root development this remarkable facet of root behaviour tends to be hidden beneath the veil of statistical averaging. Progress in other fields is providing intriguing insights into the phenomenon of developmental instability, how it is generated at the molecular and cellular levels and the genetic mechanisms by which it is buffered. This review will consider the existence of developmental instability in roots, its underlying causes, its effects on root architecture, and the evidence that it is under genetic control. The hypothesis will be advanced that developmental instability in roots is an adaptive trait, and its potential relevance to root function will be discussed in both an ecological and an agronomic context.
Introduction
A root system's ability to perform its key roles, the capture of water and nutrients from the soil and providing anchorage for the shoot, is strongly dependent on its root architecture, i.e. its spatial distribution within the soil. Agriculture in the 21st century is predicted to become more and more limited by the availability and the cost of water and nutrients (Tilman et al., 2002; Lal, 2007; Pretty, 2008), placing increasing urgency on the need to improve the efficiency with which root systems capture these essential resources. It is estimated that, globally, only 30–50% of the applied nitrogen fertilizer and ∼45% of the phosphorus fertilizer is taken up by crops (Tilman et al., 2002), with the losses contributing to greenhouse gas emissions and diffuse pollution of aquatic ecosystems, as well as representing enormous economic wastage. Unfortunately, root traits are notoriously difficult to select for in breeding programmes, but there is now considerable interest in the opportunities for improving crop root architecture through new approaches (de Dorlodot et al., 2007; Hirel et al., 2007; Lynch, 2007). However, for this to be an achievable goal, it is crucial that we start with a complete understanding of the complexity of the processes that contribute to building a root system.
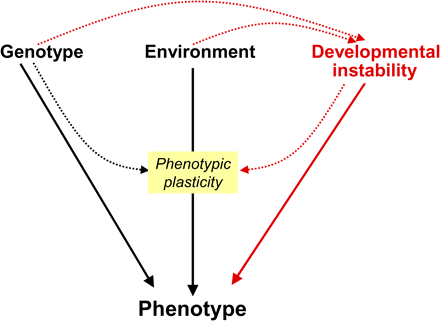
The degree to which root development is responsive to a wide range of environmental factors, including water and nutrient availability, is regarded as a prime example of the phenomenon of phenotypic plasticity (Bradshaw, 1965; Forde and Lorenzo, 2001; Malamy, 2005). Phenotypic plasticity, defined as variation that is due to environmental effects, is conventionally considered to be the only non-genetic component of phenotypic variation. However, in recent years there has been an increasing acknowledgement of an additional source of variation that arises from intrinsically stochastic processes that perturb development. This type of variation is usually termed ‘developmental instability’ and it can be considered as a third independent source of phenotypic variation, standing alongside genotype and environment (Lajus et al., 2003) (Fig. 1).
The contribution of developmental instability to phenotypic variation. Conventionally, the factors generating phenotypic variation are the genotype and the environment, with the environmental component of variation being known as phenotypic plasticity. Phenotypic plasticity also has a genetic component, because the responsiveness of an individual to the environment is dependent on the genotype (Pigliucci, 2005). However, there is an additional non-genetic factor contributing to phenotypic variation, namely developmental instability. Developmental instability causes random deviations from the target phenotype as specified by the particular combination of genotype and environment (Lajus et al., 2003). As well as affecting the phenotype directly, developmental instability can also have an indirect effect by creating random variations in phenotypic plasticity: as discussed in the text, there is good evidence that developmental instability has the potential to generate stochastic variations in the responsiveness of cells, tissues, organs, and individuals to environmental factors. In addition, there is evidence that both the genotype and the environment can modulate the degree of developmental instability in a particular character. The solid arrows indicate direct interactions with the phenotype; the broken arrows indicate indirect interactions. The section of the diagram in red represents those interactions that are not conventionally included when considering sources of phenotypic variation. See text for further information.
The purpose of this review is to examine the evidence that developmental instability is a characteristic feature of roots and one that makes a significant contribution to variation in root architecture. Although developmental instability could be viewed as a negative attribute, the case will be made that it is a potentially beneficial trait in roots (and perhaps also in other aspects of plant development) and that it could be subject to positive selection during both evolution and in breeding programmes.
Developmental instability in roots
Although not frequently commented on, it has been recognized for some time that there is a major component of root behaviour, particularly lateral root (LR) behaviour, that is non-genetic and also has no apparent environmental cause. The unpredictable nature of LR growth becomes most evident when the behaviour of individual LRs in genetically identical plants is analysed and it has been observed in a diverse range of species (May et al., 1967; Pagès et al., 1993; Pagès and Pellerin, 1994; Pagès, 1995; Zhang et al., 1999; Lecompte et al., 2001; Freixes et al., 2002; Lecompte and Pagès, 2007). Where the growth rates of individual LRs were reported, there is evidence of a 5–10-fold variation (Pagès, 1995; Freixes et al., 2002). To put the magnitude of this variation into perspective, one of the strongest environmental signals stimulating LR growth in arabidopsis (Arabidopsis thaliana L.) is nitrate, which elicits a 2-fold increase in the mean elongation rate (Zhang et al., 1999). It therefore becomes difficult to argue that environmental factors could be wholly, or even mainly, responsible for the observed variation in LR growth rates under controlled environmental conditions.
To illustrate the variability in LR growth more clearly, Fig. 2 shows the results of an experiment in which arabidopsis seedlings were grown on vertical agar plates in continuous light on two different N sources (nitrate or glutamine). The individual LR lengths from each set of seedlings have been plotted against the age of the primary root from which they emerged. Since LR initiation in arabidopsis occurs in strictly acropetal sequence (Dubrovsky et al., 2006) this allows a direct comparison between first-order LRs of similar age, correcting for variations in primary root growth between seedlings. Within each age group, and under both N conditions, it can be seen that there is a wide range of LR lengths and that, for the oldest LRs (7–8 d), lengths differed by up to 10-fold. Furthermore, most of this variation occurred within individual seedlings, again indicating that environmental effects are not primarily responsible.
Variation in LR lengths along the primary root axis of Arabidopsis thaliana L. seedlings. Seed (Col-0) was germinated and grown on vertical agar plates containing either 1 mM KNO3 or 0.5 mM glutamine at 24 °C as previously described by Zhang and Forde (1998). Four-day-old seedlings were transferred from the germination plates to fresh plates (three seedlings per 10 cm square plate, three plates per treatment) containing the same N sources. Root lengths were measured after a further 7 d growth. To correct for differences in primary root growth rate between seedlings, the LR lengths have been plotted against their age rather than the distance from the base of the hypocotyl. Their age was estimated by following the growth of the primary root at 24 h intervals and calculating the time at which the primary root tip passed the point at which the LR subsequently emerged. [Note that this will consistently overestimate the actual age of the LRs by ∼8 h, since it has been observed that the first mitotic divisions preceding formation of a LRP occur around 3 mm behind the root tip in arabidopsis (Dubrovsky et al., 2001) and allowing for a mean primary root growth rate in this experiment of 0.4 mm h−1]. The vertical line through the centre of the x-axis represents the primary root and the data for the nitrate-grown seedlings have been plotted on the left and those for the glutamine-grown seedlings on the right. Different symbols represent data from separate seedlings. The continuous red lines indicate the mean LR lengths along the primary root axis for each treatment. To obtain these, the LR lengths in 12 intervals, each of 10 h, were pooled, their means calculated, and a curve was hand-drawn through the resulting points (not shown). The dashed black lines indicate the apparent maximum length achievable by a LR of a given age under these growth conditions. Note that because of gravitropic curvature of the LRs, the outer limits of the volume of soil explored by the root system will be less than this would imply.
Lateral root growth is a complex developmental process and so stochastic processes can intervene at more than one stage to create the variation in LR length. In arabidopsis, the stages where there is evidence of stochasticity are the duration of early LR development (between initiation and emergence) (Dubrovsky et al., 2006), the duration of growth (Al-Ghazi et al., 2003), and the growth rate (Freixes et al., 2002). In the case of the experiment in Fig. 2, it was the last of these three factors that seemed to be the major contributor to the variation in LR lengths (data not shown). For example, when the growth rates were compared between LRs in their 4th day after emergence, there was an average 3-fold difference between the fastest and the slowest growing LR within an individual glutamine-grown seedling. In addition, there were also marked differences between LRs in their growth kinetics, with a proportion of LRs showing growth rates that plateaued or even declined after the 4th day. Other studies have found similar variations in the trajectory of growth rate during the lifetime of an individual LR in different species (Pagès, 1995; Lecompte et al., 2001; Freixes et al., 2002).
There are a number of other aspects of root development that show evidence of developmental instability. For example, variations in root hair density have been reported between genetically identical arabidopsis seedlings growing under controlled conditions, a phenomenon that was accentuated under S, N or K deficiencies (He et al., 2004). A study of the spacing between LRs on the arabidopsis primary root found that although the mean inter-LR distance appears to be genetically determined (in that there were consistent differences between arabidopsis accessions), there was a 25-fold difference between the maximum and minimum inter-LR distance in individual pericycle cell files (Dubrovsky et al., 2006). Significantly, there appears to be no correlation between the growth rate of a LR and its distance from the adjacent laterals (Draye, 2002; Dubrovsky et al., 2006), indicating that variation in spacing is not contributing to the variability in LR growth.
It is also necessary to consider the extent to which developmental instability at the physiological level could indirectly influence root architecture by creating non-genetic variation in phenotypic plasticity. For example, wide variations between individual roots in their trajectories after gravistimulation have been reported (reviewed by Trewavas, 2003). If, as seems likely, this variability in responsiveness extends to other environmental stimuli, it adds another dimension to the complexity of non-genetic variation. It implies that an additional component of non-genetic variation in root architecture (within and between individuals) arises from the interaction between environmental variation and non-genetic variation in phenotypic plasticity (due to the effects of developmental instability).
Because so little attention has been paid to developmental instability in roots, a brief diversion will be taken to explore the large body of knowledge concerning this phenomenon in other biological systems, its origins and the evidence that it is genetically controlled, and its potential relevance as an evolutionarily relevant trait.
Developmental instability through the ages
In The Origin of Species, Darwin stated his view that ‘any variation which is not inherited is unimportant for us’ (Darwin, 1859). Despite this early setback, there has been a long and distinguished history of investigations into phenotypic variation that has no apparent genetic or environmental cause. In the earliest study of its kind, Pearson et al. (1901) analysed 22 different structures in plants and fungi, including veins and spines on leaves, leaf indices, and seed pods. Pearson noted that these exhibited ‘a certain degree of variation combined with a certain degree of likeness’. Later Astauroff studied a range of characters in different animal species and found evidence of developmental variation that could not be explained by either environmental or genotypic effects (Astauroff, 1930). By the 1950s this phenomenon was attracting the interest of evolutionary biologists (Mather, 1953; Sakai and Shimamoto, 1965), and it was then that it first became referred to as developmental instability.
The overwhelming majority of studies on developmental instability has focused on animals rather than plants, despite the latter's suitability for this kind of investigation. In plants, genetically identical individuals can easily be propagated and the modular pattern of plant development also allows comparison of repeated structures within a single individual. In one particularly notable botanical study, Roy made hundreds of thousands of observations on petal number as well as thousands of observations on leaf tooth number in Nyctanthes arbor-tristis (Roy, 1962). His meticulous analysis, carried out over several months in different seasons, revealed a high degree of variability in both traits, both within and between individual trees. In the case of petal number, where his data were most complete, he felt able to conclude that environmental effects could not account for this variability.
Early evidence that developmental instability in plants and animals is genetically controlled, and therefore potentially subject to natural selection, came from studies of asymmetry of sternopleural chaetae number in Drosophila melanogaster (Mather, 1953) and variability in floral and foliar morphology in Nicotiana rustica (Paxman, 1956). Paxman observed within-plant instability in two floral traits (pistil and stamen length) and one vegetative trait (leaf shape) and found that the degree of instability differed between different N. rustica genotypes. Later studies with other plant species confirmed that different genotypes generally display different levels of variability in developmental traits (Williams, 1960; Roy, 1962; Sakai and Shimamoto, 1965). Two recent studies using recombinant inbred lines of arabidopsis have mapped specific quantitative trait loci (QTL) that affect developmental instability in above-ground morphological traits (Hall et al., 2007; Sangster et al., 2008).
The same studies also revealed another important facet of developmental instability by showing that the degree of instability is not a global phenomenon for the whole organism but can vary independently in different units of development. For example, in two species of Nicotiana it was found that instability of floral development was inherited independently of foliar instability (Paxman, 1956; Sakai and Shimamoto, 1965) and that while there seemed to be a common mechanism controlling instability in the two floral characters, flower and leaf stability were independent. A meta-analysis of data sets from 11 invertebrate species similarly concluded that the genetic basis of developmental stability is character-specific (Clarke, 1998). It therefore seems safe to conclude that developmental instability can operate differentially in different developmental processes within a single organism.
Thus it can be seen that there is good evidence for the existence of a source of phenotypic variation in plants that is independent of genetic and environmental effects. Furthermore, the degree to which a particular character displays developmental instability has been shown to be, at least partly, under genetic control. In the following two sections the significant progress that has recently been made in understanding the processes that generate developmental instability, the mechanisms that act to buffer development against these stochastic processes, and the genetic basis for these opposing forces will be considered.
Origins of developmental noise
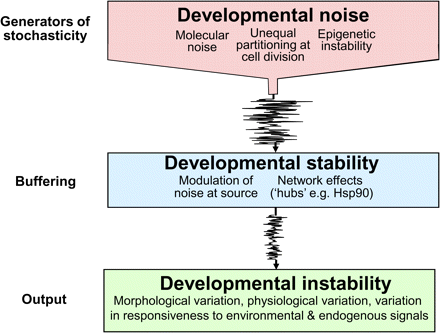
It is important to be clear about the terminology employed in the following discussion because different usages are in circulation. Here we will use the term developmental noise to refer to the stochastic processes that cause a developing trait to deviate from its expected path under given genotypic and environmental conditions (Van Dongen and Lens, 2002). Developmental stability is the buffering capacity that counteracts the phenotypic effects of developmental noise and developmental instability is the residual non-genetic variation that is resistant to buffering (Fig. 3). Microenvironmental effects are those originating from environmental variations (internal or external to the plant) that occur even when the macroenvironment is controlled.
The interaction between the processes that generate developmental instability and those that buffer it. See text for further details.
In studies of multicellular organisms it will usually be difficult to discern the boundary between phenotypic variation resulting from intrinsic stochastic processes and that due to ‘microenvironmental’ effects of unknown origin (Simons and Johnston, 2006). Microenvironmental factors would include such things as minor variations in light intensity or temperature occurring within a growth chamber, differential shading of the upper and lower leaves within a plant, or heterogeneity in the internal environment of the organism. When environmental gradients occur on a small scale they cannot be readily quantified and their contribution to stochastic developmental behaviour may be difficult to assess. However, there may be ways to obtain some indication of the degree to which variation in a trait is due to developmental noise or to microenvironmental effects. For example, one could compare the coefficient of variation (CV) for the trait amongst groups of plants grown under conditions where microenvironmental effects are minimized (e.g. within the same Petri dish) with that for groups of plants grown under controlled, but less uniform conditions (e.g. in different parts of the same growth chamber). The expectation would be that where stochastic processes are largely at work, the CVs would be similar, whereas if microenvironmental effects predominate the CV would be significantly greater in the second case. Another indication that microenvironmental effects are insufficient to account for variation in a trait would be where the magnitude of the variation observed in a controlled environment is similar to or greater than the magnitude of the response induced by macroenvironmental factors (as was noted above in the case of the LR growth rates). A contrasting example where microenvironmental factors were found to be the predominant cause of within-plant variation comes from studies of rhizome lengths in Carex spp. (de Kroon et al., 1994). Here the variation in rhizome length could largely be accounted for by the variation in light intensity caused by the variable orientation of the lateral buds in relation to the soil surface.
In some respects, it might not always be important to know whether the variation in a particular trait is due to microenvironmental effects or to developmental noise since the outcome in terms of the effect on the fitness of the individual or the population may be indistinguishable. Furthermore, it will be seen that even the processes that act to buffer the two sources of non-genetic variation may be related.
If one considers events at the molecular level it is not difficult to identify one of the most important underlying causes of developmental noise. Biochemical reactions are inherently stochastic because they depend on random intermolecular collisions (Shahrezaei and Swain, 2008). In general, this stochasticity is greatest when the numbers of molecules involved are low, because it is then that the randomness of individual reaction events will lead to significant statistical fluctuations in molecule numbers and reaction rates. Experiments using single-cell reporter assays have made it possible to observe dynamic changes in gene expression in individual bacterial cells (Elowitz et al., 2002; Swain et al., 2002). These studies confirmed the expectation that expression at the single gene level is extremely stochastic and demonstrated that there are two components to this noise: intrinsic and extrinsic. For an individual gene, intrinsic noise is due to stochastic events in the process of its own expression, while extrinsic noise is the component that is imposed on the gene by fluctuations in the abundance of other cell components that have a global effect on gene expression in that cell. Extrinsic noise can have its origins in the heterogeneity of any of a variety of factors that affect gene expression, including heterogeneity at the cellular level (e.g. in cell size or in cell cycle stage) or in upstream signal transduction (Raser and O'Shea, 2005). For example, stochasticity in the expression of a transcription factor (or regulatory RNA molecule) can propagate stochasticity to the genes that it regulates, an effect that is amplified if there is a linear cascade of transcription factors (Hooshangi et al., 2005). Other biochemical processes influenced by noise include ion-channel gating, cytoskeleton dynamics, and protein translocation across membranes (Rao et al., 2002).
To add further to the sources of developmental noise, there are also processes that generate stochastic differences between a mother cell and its mitotically-derived daughter cells. For example, when cell division occurs, most of the cytoplasmic components are partitioned randomly between the two daughter cells. Where molecules or organelles are present in small numbers in the cell this creates considerable potential for random divergence between the daughter cells (McAdams and Arkin, 1999). Furthermore, there is also the potential for developmental noise to be generated through spontaneous epigenetic changes in somatic tissues (Blewitt et al., 2004). This would involve localized alterations in the chromatin structure or in the DNA methylation state that can potentially affect gene expression. Although the epigenetic state is considered to be mitotically heritable, studies with mammalian cell cultures have indicated that the DNA methylation state at an individual locus may be inherited with an efficiency as low as 94% (Wigler et al., 1981). Consistent with this, there are reports of cell-to-cell differences in the epigenetic state of many alleles within the same mammalian tissue and, at least in some cases, this is reflected at the level of gene expression (Blewitt et al., 2004). The extent to which this instability exists in plants is less clear, but genetic studies in several species have revealed a number of examples of metastable epialleles that generate somatic mosaics or chimaeric plants (Cubas et al., 1999; Miura et al., 2009; Sekhon and Chopra, 2009), indicating the potential for spontaneous epimutations to contribute to the generation of developmental noise in plants as well.
Once the extent to which gene expression and other cellular processes are inherently stochastic is appreciated, it can be understood how it can be that each individual in a genetically homogeneous population of cells or organisms is unique. This phenomenon, which is referred to as ‘non-genetic individuality’, was first discovered in the 1970s through studies of the variability of the chemotactic response in cells of a clonal bacterial population (Spudich and Koshland, 1976). Individuality has also been discussed in the context of plants with respect to the non-genetic differences that are seen between cells and tissues in their sensitivity to hormones such as auxin, ethylene, and gibberellin (Trewavas, 1999; Gilroy and Trewavas, 2001). These differences in sensitivity are exactly what one would predict as a consequence of stochasticity at the molecular level, which could readily generate variations in the concentrations of key regulatory molecules.
The individuality in the behaviour of cells during the development of a multicellular organism could also scale up to produce significant variation at the organ and whole organism level. For example, in plants, cell-to-cell variations in hormone sensitivity could have a major effect on how a nascent organ or tissue, consisting of a relatively small number of cells, responds to developmental signals. Small differences between cells at the beginning of a developmental process could be transmitted to their daughter cells, and supplemented over time by further stochastic events, could lead to significant phenotypic divergence between genetically identical individuals or repeating units within an individual. These sources of developmental noise, perhaps combined with additional microenvironmental effects, would seem to provide an adequate explanation for the non-genetic variation in floral and foliar traits that was the subject of such painstaking analysis in the 1950s and 1960s (Went, 1953; Paxman, 1956; Roy, 1962; Sakai and Shimamoto, 1965).
Keeping the noise down
Considering the intrinsic stochasticity of the processes that underlie development one might wonder why development is not even more unstable than it is. In recent years considerable advances have been made towards understanding the mechanisms that exist to buffer development against perturbation of different kinds. Progress has come from work on two related topics, developmental stability (buffering stochastic processes) and ‘canalization’ (Waddington, 1942), the process that buffers development against perturbation by genetic and environmental factors. The precise relationship between developmental stability and canalization is controversial (Debat and David, 2002), but recent evidence suggests at least some overlap between the two buffering systems (Salathia and Queitsch, 2007).
For those working on canalization, interest in the last decade has focused on the molecular chaperone Hsp90 after it was reported to act as a phenotypic capacitor in D. melanogaster (Rutherford and Lindquist, 1998). Disruption of Hsp90 function has been shown to lead to changes in a wide variety of morphological phenotypes in both arabidopsis and D. melanogaster, apparently as a result of exposing cryptic genetic variation (Rutherford and Lindquist, 1998; Queitsch et al., 2002; Sangster et al., 2007, 2008). It has been suggested that Hsp90’s canalizing function is a by-product of its central role in stabilizing the conformation of metastable regulatory proteins, including diverse regulators of growth and development (Queitsch et al., 2002).
There is evidence that Hsp90 also buffers developmental noise. Studies with arabidopsis (Sangster et al., 2007) and D. melanogaster (Queitsch et al., 2002; Milton et al., 2006) have reported that interfering with Hsp90 function leads to increased phenotypic variability in the absence of genetic variation. However, in D.melanogaster not all traits are apparently subject to Hsp90 buffering, its reported effects being largely specific to those quantitative traits that are normally most invariant (‘threshold’ traits) (Milton et al., 2006).
Hsp90 is just one of a large group of molecular chaperones that serve many functions under both normal and stressful conditions (Nollen and Morimoto, 2002). Each of these chaperones typically engages in low-affinity, dynamic interactions (‘weak links’) with many other proteins, and as such they behave like hubs in cellular networks (Soti et al., 2005). Simulation modelling has shown that scale-free networks, such as cells or the Internet, are inherently robust even to high failure rates at individual nodes, but are vulnerable to attack or perturbation at network hubs (Albert et al., 2000). This prediction has been confirmed by a large-scale deletion analysis in yeast which showed that >60% of proteins with more than 15 network links were essential, compared to ∼10% of proteins with fewer than five links (Jeong et al., 2000). This network model view fits with the idea that canalization is simply the inevitable consequence of the evolution of complex regulatory networks (Siegal and Bergman, 2002) and helps us to understand the reasons for Hsp90’s importance as a phenotypic capacitor. It also leads to the prediction that other highly connected proteins could have a similar role in buffering development against genetic, environmental, and stochastic perturbations (Sangster et al., 2004). In confirmation of this, a detailed study of genetic interactions in Caenorhabditis elegans identified a class of ‘hub’ genes which encode chromatin regulators and which have a conserved role as phenotypic capacitors across the animal kingdom (Lehner et al., 2006). Similar work with yeast revealed a set of 300 genes (5% of the genome) that fulfil the role of phenotypic capacitors (Levy and Siegal, 2008).
Mechanisms that operate to control the amount of stochasticity in gene expression do not act exclusively by buffering noise after it has been produced. Evidence that gene expression noise can be controlled at its source comes from studies showing that essential genes typically have lower noise levels than dispensable genes (Kaufmann and van Oudenaarden, 2007). One way in which this control is exerted may be by balancing the relative rates of transcription and translation (Thattai and van Oudenaarden, 2001; Ozbudak et al., 2002). Other work has demonstrated how different promoter sequences can generate the same mean mRNA abundance but with very different noise characteristics (Raser and O'Shea, 2004), and there is a growing list of genetic factors shown to contribute to the generation of gene expression noise (Ansel et al., 2008).
When noise can be a good thing
It seems natural to think of developmental instability as a universally undesirable trait, one that simply indicates a failure of the processes that buffer developmental noise, leading to a departure from the ideal phenotype. Thus developmental instability, exhibited for example as a breakdown in bilateral symmetry, is often considered an indicator of environmental or genetic stress (Parsons, 1992). Nevertheless, there is an impressive amount of theoretical and experimental evidence behind the idea that, under some circumstances, there can be a selective advantage to the increased phenotypic variability that comes with a relaxation of developmental stability (or increase in developmental noise).
For microbial populations, it is already well established that stochastic processes (such as gene expression noise) play a fundamental role in generating phenotypic diversity and in doing so are able to confer a fitness advantage on the population as a whole (Fraser and Kaern, 2009). An example is the phenomenon of bacterial persistence where a small subset of cells spontaneously enters into a dormant state in which they can survive adverse conditions to reseed the population when conditions improve (Lewis, 2007). This is an example of a strategy known as diversified bet-hedging, which is a common application of stochasticity in micro-organisms (Fraser and Kaern, 2009). Diversified bet-hedging is where the risks associated with living in a variable environment are spread by means of phenotypic variance within a single genotype. The idea can be summarized by the adage of not putting all your eggs in one basket. The archetypal example of bet-hedging is seed germination (Cohen, 1966). In a constantly favourable environment, alleles determining immediate germination would be selected, whereas in an unpredictable environment, alleles that cause germination to be spread over an extended period of time will be favoured (Evans and Dennehy, 2005). Stochasticity in the timing of germination ensures that at least a fraction of the progeny will survive to reproduce. Other examples where developmental instability in plants could be selected for as part of a bet-hedging strategy include flowering time, where asynchrony favours outcrossing and prolongs the flowering period (Normand et al., 2002) and bud dormancy, where variability is predicted to be beneficial if there is year-to-year variation in the risk of herbivory (Nilsson et al., 1996).
Sources of developmental instability in lateral roots
Returning to roots, what can we now say about the likely reasons for the variability in LR development? It seems very likely that much of the observed variation can be traced back to the earliest stages of LR formation when stochastic events at the molecular and cellular level would have a lasting influence on subsequent developmental processes.
As is well known, LRs arise post-embryonically from pericycle cells known as founder cells a short distance behind the apical meristem of the primary root (Malamy and Benfey, 1997; De Smet et al., 2006). Formation of the LR begins when the founder cells undergo a series of asymmetric anticlinal divisions, immediately followed by periclinal divisions to form the dome of dividing cells that is the LR primordium. Significantly, there appears to be no agreement about the number of founder cells that contribute to LR initiation in any species. In arabidopsis, one estimate put the average number at around 11 (Laskowski et al., 1995), but it was noted that there was considerable variation in the longitudinal and radial dimensions of the early LR primordium, indicating significant deviations from that number. A more recent study found that there were two distinct morphogenetic types of LR initiation in arabidopsis and estimated that the minimum number of founder cells for the ‘longitudinal unicellular’ type was three, but that additional cells could be recruited (Dubrovsky et al., 2001). In the case of the other type (‘longitudinal bicellular’), the estimated minimum number was twice this figure. The evidence therefore suggests that there can be unpredictability even in so fundamental an event in the life history of a LR as the number of founder cells that establish it.
Hormonal signalling is known to be important for LR initiation, with auxin playing a dominant role (De Smet et al., 2006). Thus stochastic differences between pericycle cells (i.e. ‘non-genetic individuality’) in their sensitivity to these signals could be one factor that influences the number of founder cells. Experimental evidence indicating a stochastic distribution of auxin sensitivities in the pericycle cell population has been reviewed before (Gilroy and Trewavas, 2001). Additional scope for stochasticity in the crucial early stages of LR development would come during cell division with the potentially unequal partitioning of cytoplasmic components between daughter cells (McAdams and Arkin, 1999).
Cytological evidence for developmental instability during early LR development comes from the observation that the number of cortical and endodermal cell files in the mature LR is variable (7–11 and 7–10, respectively), whereas in the primary root they are fixed at 8 (Dolan et al., 1993). It has also been observed in a number of plant species that the apical diameter of secondary and higher order roots is highly variable within each order, and that it is broadly correlated with the growth rate (Drew et al., 1973; Cahn et al., 1989; Lecompte and Pagès, 2007). It is not clear what relationship there is, if any, between the apical diameter and the number of cortical and epidermal cells files, or between the variability in these parameters and the variability in the LR growth rate. However, based on an observed highly significant correlation between the early growth rate and the final LR length, it has been suggested that the growth potential of a LR is determined early in development (Pagès, 1995). It will be interesting to establish whether there are cytological events during the formation of the LR meristem that establish both its morphology and its future proliferative potential. Table 1 provides a summary of evidence for variability in events and processes that could contribute to the developmental instability associated with LRs.
Sources of developmental instability affecting root architecture
| Evidence of stochasticity | Effects on root architecture | |
| Cellular | Auxin sensitivity of pericycle cells1 | Via effect on inter-LR spacing and no. of founder cells? |
| Cytological | No. of LR founder cells2,3 | Possibly related to apical diameter of LRs and meristem size |
| No. of cortical and endodermal cell files in LRs4 | ||
| Developmental | Apical diameter of LRs (correlated with growth rate)5–7 | Variation in specific root length |
| Time to LR emergence8 | Variation in LR length | |
| LR growth rate/kinetics9–11 | ||
| Duration of LR growth12 | ||
| Inter-LR spacing8 | Variation in LR density | |
| Root hair density13 | Variation in root hair density | |
| Physiological | Gravitropic response14 | Variation in growth angle |
| Evidence of stochasticity | Effects on root architecture | |
| Cellular | Auxin sensitivity of pericycle cells1 | Via effect on inter-LR spacing and no. of founder cells? |
| Cytological | No. of LR founder cells2,3 | Possibly related to apical diameter of LRs and meristem size |
| No. of cortical and endodermal cell files in LRs4 | ||
| Developmental | Apical diameter of LRs (correlated with growth rate)5–7 | Variation in specific root length |
| Time to LR emergence8 | Variation in LR length | |
| LR growth rate/kinetics9–11 | ||
| Duration of LR growth12 | ||
| Inter-LR spacing8 | Variation in LR density | |
| Root hair density13 | Variation in root hair density | |
| Physiological | Gravitropic response14 | Variation in growth angle |
References: 1. Gilroy and Trewavas (2001); 2. Laskowski et al. (1995); 3. Dubrovsky et al. (2001); 4. Dolan et al. (1993; 5. Drew et al. (1973); 6. Cahn et al. (1989); 7. Lecompte et al. (2007) ; 8. Dubrovsky et al. (2006); 9. Pagès (1995); 10. Lecompte et al. (2001); 11. Freixes et al. (2002); 12. Al-Ghazi et al. (2003); 13. He et al. (2004); 14. Trewavas (2003)
Sources of developmental instability affecting root architecture
| Evidence of stochasticity | Effects on root architecture | |
| Cellular | Auxin sensitivity of pericycle cells1 | Via effect on inter-LR spacing and no. of founder cells? |
| Cytological | No. of LR founder cells2,3 | Possibly related to apical diameter of LRs and meristem size |
| No. of cortical and endodermal cell files in LRs4 | ||
| Developmental | Apical diameter of LRs (correlated with growth rate)5–7 | Variation in specific root length |
| Time to LR emergence8 | Variation in LR length | |
| LR growth rate/kinetics9–11 | ||
| Duration of LR growth12 | ||
| Inter-LR spacing8 | Variation in LR density | |
| Root hair density13 | Variation in root hair density | |
| Physiological | Gravitropic response14 | Variation in growth angle |
| Evidence of stochasticity | Effects on root architecture | |
| Cellular | Auxin sensitivity of pericycle cells1 | Via effect on inter-LR spacing and no. of founder cells? |
| Cytological | No. of LR founder cells2,3 | Possibly related to apical diameter of LRs and meristem size |
| No. of cortical and endodermal cell files in LRs4 | ||
| Developmental | Apical diameter of LRs (correlated with growth rate)5–7 | Variation in specific root length |
| Time to LR emergence8 | Variation in LR length | |
| LR growth rate/kinetics9–11 | ||
| Duration of LR growth12 | ||
| Inter-LR spacing8 | Variation in LR density | |
| Root hair density13 | Variation in root hair density | |
| Physiological | Gravitropic response14 | Variation in growth angle |
References: 1. Gilroy and Trewavas (2001); 2. Laskowski et al. (1995); 3. Dubrovsky et al. (2001); 4. Dolan et al. (1993; 5. Drew et al. (1973); 6. Cahn et al. (1989); 7. Lecompte et al. (2007) ; 8. Dubrovsky et al. (2006); 9. Pagès (1995); 10. Lecompte et al. (2001); 11. Freixes et al. (2002); 12. Al-Ghazi et al. (2003); 13. He et al. (2004); 14. Trewavas (2003)
Is developmental instability in roots under genetic control?
Given the potential for stochastic processes to influence the early development of the LR, it might be argued that the highly variable nature of LR development is no more than an unavoidable consequence of the post-embryonic nature of the process. A strong counter-argument to this view comes from consideration of the development of the cluster (or proteoid) roots that are produced by some plant species as an adaptation to nutrient-poor soils (Watt and Evans, 1999a). These determinate LRs (or ‘rootlets’) initiate spontaneously at high density on the parent root and their subsequent development, elongation, and growth cessation is highly synchronous (e.g. see Fig. 3b in Watt and Evans, 1999b), culminating in an ‘exudative burst’ when a large-scale efflux of enzymes and organic acids occurs simultaneously in the hundreds of rootlets within a single cluster. The highly choreographed nature of this process suggests that where stochasticity of root development would be maladaptive it can be buffered extremely effectively.
The remarkable contrast between the predictability of the behaviour of cluster roots and the stochastic behaviour of most other kinds of LRs is one indication that developmental instability in LRs is a trait that can be genetically regulated. As already discussed, there is already good evidence from studies of non-genetic variation in above-ground traits that developmental instability in plants is under genetic control, making it likely that similar mechanisms will operate below ground. However, the paucity of data on developmental instability in roots means that evidence for natural variation in this trait is less easy to find. Examination of published data where different root parameters, such as inter-LR spacing (Draye, 2002), primary root growth (Beemster et al., 2002), and total root length (Armengaud et al., 2009), have been measured in different accessions or cultivars suggests that there are genotypic differences in the variance of these traits. Analysis of within-plant variance in LR growth rates in arabidopsis has indicated the existence of differences between accessions (T Remans and BG Forde, unpublished results). Other evidence comes from studying the effect of chemical impairment of Hsp90 activity in three different populations of recombinant inbred lines of arabidopsis (Sangster et al., 2008). The treatment resulted in increased variability in both hypocotyl and primary root lengths, leading the authors to conclude that developmental stability in these parameters is a phenotypic trait that can be affected by natural variation.
The availability of extensive collections of accessions, recombinant inbred lines and near-isogenic lines in some model plant species, particularly arabidopsis, should make it possible to map QTLs for developmental instability in root development and to identify the genes involved. At the same time, the QTL approach should reveal whether developmental instability is controlled separately at different steps in root development. In the shorter term, it will be interesting to establish whether Hsp90’s role as a phenotypic capacitor extends to one or more of the stages of LR growth and development.
Could developmental instability be beneficial for root architecture?
For developmental instability to be relevant in an evolutionary sense there must not only be evidence of heritable differences in the degree of natural variation in the trait, there must also be evidence that it has an effect on the fitness of the individual or the population. We have seen how, from bacteria to plants, there are clear examples showing how it is possible for developmental instability in other traits to be advantageous. What then might be the benefits of developmental instability in roots?
Below, four possibilities are speculatively advanced, two of which (2 and 4) relate to the effects of developmental instability on the architecture of an individual root system, and two (1 and 3) that apply to situations where there is significant, non-genetic variation in root architecture between plants. Although this review has focused primarily on the variation between repeated modules within a single plant, the developmental instability responsible for this variation inevitably generates variation between plants as well. For example, in the experiment described in Fig. 2 (and in other similar experiments), it was observed that the CV within treatments was consistently much higher for total LR length and LR number per seedling than for primary root length, apparently reflecting the greater developmental instability associated with LRs compared with primary roots. This makes it reasonable to consider the ecological implications of non-genetic variation occurring both within and between plants.
Note that the four possibilities outlined here are not mutually exclusive, but it is envisaged that their adaptive advantages would apply differentially depending on the ecology and life history of the species in question.
A bet-hedging strategy
Where developmental instability in other systems is used as the driving force for a bet-hedging strategy, it is seen as an adaptation to the inherent unpredictability of natural environments (Simons, 2009). In the case of roots, it is clear that the environment they are called upon to inhabit and explore is exceptionally stochastic, one in which random spatial and temporal heterogeneities in the supply of water and nutrients are the norm. Although phenotypic plasticity is one important mechanism for adapting to these kinds of heterogeneity (Magyar et al., 2007), it suffers the inevitable disadvantage that it is unable to predict future stochastic events. Some stochasticity in the process of root development would provide a means of conferring a root architecture on some individuals that by chance was ideally suited to the soil conditions that they subsequently experienced. This would be analogous to the strategy whereby an individual plant produces a heterogeneous population of seeds differing in their germination time and dispersion potential as a way of coping with spatiotemporal variability in the habitat (Matilla et al., 2005). In this scenario, alleles promoting diversity in root architecture amongst an individual's progeny would be favoured over alleles promoting uniformity.
As part of a foraging strategy
The same problem of how plants cope with a stochastic environment can also be considered at the level of the individual. Until a root system encounters a resource-rich patch it can have no way of ‘knowing’ where that patch is located. A stochastic element to the expansion of the root system could therefore be the most efficient means of sampling the soil environment for the presence of a patch. Once the patch has been detected by a root tip, the appropriate response can be triggered, i.e. increased proliferation of roots within the patch (Hodge, 2006). This could be thought of as analogous to the ‘random walk’ strategy used by animals to find resource-rich patches (or prey) when they have no a priori knowledge of the location of their target (Bartumeus et al., 2005). The case has been made for the usefulness of the Marginal Value Theorem of animal foraging behaviour (Charnov, 1976) in understanding foraging behaviour by roots (McNickle and Cahill, 2009). However, such classical optimality-based models assume that the animal has complete knowledge of the spatiotemporal distribution of resources (Stephens and Krebs, 1986), a concept that cannot readily be applied to plants. Therefore, while the Marginal Value Theorem may be appropriate for modelling root behaviour within a patch (McNickle and Cahill, 2009), other approaches are needed when considering how a root system optimizes its chances of locating the patch in the first instance. The complications of applying animal foraging models such as the Marginal Value Theorem to plants have been discussed elsewhere (Hutchings and de Kroon, 1994). A computational modelling approach could provide an excellent means of testing the hypothesis that some degree of stochasticity in root development may be optimal for locating resource-rich patches in a stochastic environment.
Reducing underground competition between genetically related neighbours
Inter-specific variation in root architecture has been identified as a strategy to reduce niche overlap and therefore competition between species (Parrish and Bazzaz, 1976). For closely related plants of the same species growing in close proximity, a way of achieving the same outcome may be through the generation of stochastic differences in root architecture between individual plants. If this were the sole driving force for developmental instability in roots, it would lead to the prediction that non-genetic variation in root architecture between individuals would be most prevalent in species where genetically identical or closely related individuals are frequently in competition (such as those reproducing clonally), or where intra-specific genetic variation in root architecture is minimal.
Increasing the efficiency of soil exploration at the single plant level
The idea has already been put forward that the non-genetic variability in LR growth enables a root system to explore a greater volume of soil than if the same total root length were produced deterministically (Pagès et al., 1993). This effect is illustrated in Fig. 2 by considering the composite data from the multiple seedlings as a model for a single larger root system. It can be seen that the volume of soil that could potentially be explored through a stochastic pattern of growth (indicated by the dotted black lines) is much greater than the volume that would be explored if growth were deterministic and all the LRs were of the average length (continuous red lines). In this scenario, an ability to accentuate the stochasticity of LR growth under nutrient-limiting conditions would have a similar effect as the ability to increase specific root length. The latter is a well-known example of root plasticity that serves to minimize the costs of constructing a root system while maintaining the ability to explore the soil as effectively as possible (Forde and Lorenzo, 2001). Another potentially beneficial property of a stochastically generated root system is the randomizing effect that it has on the distribution of the root tips within the soil. The significance of this lies in the pre-eminent contribution that root tips make to many of the most important aspects of root function. Thus the apical region of the root is an important site for the uptake of less mobile nutrients (such as Pi or Fe) because these become depleted behind the growing root tip and because the root tip is the location of the root hair zone and a hot-spot for exudations that mobilize these nutrients (Vance et al., 2003). Consequently, an optimal deployment of root tips within the soil would both minimisze competition between root tips for soil resources and maximize the efficiency of exploration of the entire soil volume, traits that may be enhanced by an element of stochasticity in LR development. Root systems also have another important role as information-gathering devices and it is the root tip that is the primary site for the sensing of a multitude of environmental signals (Darwin, 1880; Takahashi and Scott, 1993; Siebrecht et al., 1995; Aiken and Smucker, 1996; Svistoonoff et al., 2007; Walch-Liu and Forde, 2008). A dispersed pattern of distribution in which the root tips are located randomly throughout the occupied volume of soil, rather than only at the outer extremities of that volume (see Fig. 2), could therefore represent another potential advantage of developmental instability in roots.
Thinking agronomically
The key question from an agronomic perspective is what would be the implications of different degrees of developmental instability for the functionality of the root system of crop plants? It seems likely that intensive breeding would have selected for alleles that confer increased phenotypic stability and uniformity on the crop, at least for above-ground traits, leading to reduced levels of developmental instability in crop plants compared with their wild relatives. A comparison of developmental instability in different commercial varieties of N. tabacum found that foliar traits were much more stable in two cigar cultivars than in cultivars that had been bred for cigarette production (Sakai and Shimamoto, 1965), suggesting that the breeding programmes had inadvertently selected for different levels of developmental instability. Might differences in developmental instability for root traits similarly exist between cultivars of other crop species, and might the degree of developmental instability that exists in wild relatives be diminished in the intensively bred genotypes? These are potentially important questions because root systems represent a substantial metabolic investment by the plant, costing up to 50% of daily photosynthesis (Lynch, 2007). If, as discussed above, a less stochastic root system is less effective at exploring the soil volume for a given root length, there is likely to be a penalty in terms of crop productivity. The corollary of this argument is that there may be an optimum level of developmental instability for root architecture that could be a selection target in breeding for improved efficiency of water and nutrient capture.
Because experimental approaches to investigating the relationship between developmental instability and fitness under field conditions are likely to prove difficult, some of the crucial questions about the agronomic (or ecological) significance of variations in developmental instability may be most effectively addressed using computational modelling. Existing root architectural models often already incorporate an element of stochasticity as a way of simulating a realistic root system (Pagès, 1999). However, to take stochasticity fully into account, the model should incorporate the ability to adjust the degree of developmental instability and to examine the consequences for root architecture and root system performance (e.g. in water and nutrient uptake).
If genetic analysis shows developmental instability at different stages of root development to be inherited independently, it will be also necessary to introduce this additional layer of complexity into the computational model. For example, differences in the degree of developmental instability in the time to LR emergence may affect root architecture quite differently from, say, differences in the degree of developmental instability in the rate of LR growth. Furthermore, it will not be sufficient to consider the effect of different levels of developmental instability on the performance of the individual root system in isolation. As discussed, stochasticity may confer an advantage when roots of genetically identical or closely related plants are in direct competition (which would include a crop monoculture). Finally, to take into account the possibility of diversified bet-hedging, models will also have to consider the possibility that a certain level of developmental instability may be sub-optimal for an individual growing under average conditions, but beneficial at the population level in an unpredictable environment (Simons, 2009).
Concluding remarks
This review has made the case that the high level of stochasticity that characterizes many aspects of root development is a prime example of the phenomenon of developmental instability. While the processes that generate this developmental instability may be inevitable, to varying degrees, it has been seen that there are ways of buffering development against those disruptive processes if instability is maladaptive. It has, therefore, been proposed that developmental instability in roots may be an adaptive trait, and a number of possible ways in which it could confer an advantage in an ecological or agronomic context have been suggested.
If developmental instability is so important, it seems reasonable to ask why so little attention has been paid to it in the past. In this respect, there are some striking parallels with the status of phenotypic plasticity as a research topic prior to the 1980s. Bradshaw wrote his influential review on phenotypic plasticity in the mid-1960s (Bradshaw, 1965), but according to Schlichting (1986), work on phenotypic plasticity was largely neglected for the following 15 years because the focus of the day was on variation of genetic origin, with environmentally induced variation being looked on as something to be avoided at all costs. In the case of developmental instability, the barrier to its recognition has largely lain in the commonly held view, also expressed by Bradshaw (Bradshaw, 1965), that it is simply a sub-class of phenotypic plasticity where the environmental cause is unknown. When this perception is combined with the general tendency towards statistical averaging, it is easy to understand how developmental instability could have been so neglected, other than as an indicator of environmental stress (Van Dongen, 2006).
Given the parallels with the history of research on phenotypic plasticity, it is intriguing that the set of open questions posed by Bradshaw in 1965 are precisely those that are most apposite for developmental instability today. As later paraphrased by Schlichting (1986), and with the appropriate substitution of the latter term for the former, these questions are: What is the mechanistic basis of developmental instability? How is developmental instability in different traits related? What is the genetic control of developmental instability and can it be selected? How much genetic variability for developmental instability exists in natural populations? These questions are just beginning to be addressed in plants (Hall et al., 2007; Sangster et al., 2007; Tucic et al., 2008), but as yet almost entirely for above-ground traits and not from the standpoint that developmental instability could be an adaptive trait.
Answering these questions for roots will require a combination of empirical and theoretical approaches, but can only properly begin once it becomes more widely accepted that developmental instability is indeed a trait worthy of study in its own right. In the short term, computational modelling may offer the best prospects for advancing our understanding of the significance of developmental instability for root function. For example, how does changing the degree of instability in different developmental traits affect the exploitation efficiency of root systems of different topology? In the longer term, this work may take us to the point where we are able to exploit developmental instability as a trait for the breeding of crop plants with improved water- and nutrient-use efficiencies.
Others have persuasively argued that the tendency towards statistical averaging in plant science can often obscure important information about the variability in the behaviour of individuals (Trewavas, 1999, 2003; Amzallag, 2001). In this review the focus has been on what we may have been missing in terms of root development, but it seems likely that developmental instability could also play an important role in generating phenotypic variation of evolutionary relevance in the shoot. Ecologically important traits that may be worthy of closer examination in this respect could include plant height, stem elongation, apical dominance, bud dormancy, and the time to flowering. As with roots, the extent to which stochasticity in any of these traits is adaptive will probably depend on the species in question, its life history, and the ecological circumstances.
In a wider context, the conclusion that emerges from this review is that there is a need to reassess our concept of how plants exploit non-genetic phenotypic variation to cope with their changeable and unpredictable environment. Phenotypic plasticity enables a plant to alter its phenotype in response to a change in its current environment. Developmental instability also generates non-genetic variation, but of a non-directional (i.e. random) type that does not depend on environmental triggers. As such, developmental instability could be especially suited to buffering individual plants or plant populations against stochastic variations in their environment that cannot be perceived, either because they are in the future or because they are located beyond the current range of the plant's sensory systems.
In his most recent commentary on phenotypic plasticity, Bradshaw states that ‘genes must exist not only to determine character means, but also to determine character response, which adds interesting complexity to our ideas about evolution’ (Bradshaw, 2006). If genes also exist to determine not only the developmental instability of a character but also the non-genetic variation in the character response, then it seems that evolutionary biologists will have more complexity to deal with than they might have wished for.
Abbreviations
- CV
coefficient of variation
- LR
lateral roots
- Pi
inorganic phosphate
- QTL
quantitative trait locus
The author thanks Dr Tony Remans for the data in Fig. 2, which were obtained while he was a post-doc in the author's laboratory funded by a European Union Research Training Network grant HPRN-CT-2002-00247. The helpful and constructive comments of Professor Hans de Kroon (Radboud University Nijmegen) and a second anonymous referee are also gratefully acknowledged.



![Variation in LR lengths along the primary root axis of Arabidopsis thaliana L. seedlings. Seed (Col-0) was germinated and grown on vertical agar plates containing either 1 mM KNO3 or 0.5 mM glutamine at 24 °C as previously described by Zhang and Forde (1998). Four-day-old seedlings were transferred from the germination plates to fresh plates (three seedlings per 10 cm square plate, three plates per treatment) containing the same N sources. Root lengths were measured after a further 7 d growth. To correct for differences in primary root growth rate between seedlings, the LR lengths have been plotted against their age rather than the distance from the base of the hypocotyl. Their age was estimated by following the growth of the primary root at 24 h intervals and calculating the time at which the primary root tip passed the point at which the LR subsequently emerged. [Note that this will consistently overestimate the actual age of the LRs by ∼8 h, since it has been observed that the first mitotic divisions preceding formation of a LRP occur around 3 mm behind the root tip in arabidopsis (Dubrovsky et al., 2001) and allowing for a mean primary root growth rate in this experiment of 0.4 mm h−1]. The vertical line through the centre of the x-axis represents the primary root and the data for the nitrate-grown seedlings have been plotted on the left and those for the glutamine-grown seedlings on the right. Different symbols represent data from separate seedlings. The continuous red lines indicate the mean LR lengths along the primary root axis for each treatment. To obtain these, the LR lengths in 12 intervals, each of 10 h, were pooled, their means calculated, and a curve was hand-drawn through the resulting points (not shown). The dashed black lines indicate the apparent maximum length achievable by a LR of a given age under these growth conditions. Note that because of gravitropic curvature of the LRs, the outer limits of the volume of soil explored by the root system will be less than this would imply.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/jxb/60/14/10.1093/jxb/erp265/2/m_jexboterp265f02_3c.gif?Expires=1716489119&Signature=YRXlxiMR~iEFyGWwXjfISNzvvB54764qI5z1G0xxCSh8Q7ldp2lL5qlXwdteR68AS~HEU5g12nV1c44IiODi62DM8uhBjDR~eZgEVgvbQ0XGmz-PxSoaYXlRHjJKyKTaDWIvJ4Ia2ysifi88mqesweWkv8Lc6WOngi~aMbmlSRLxxTFaA7YPIZjcwt0MrHmYr7AvGCQV9oYeWThnjCLVlkBEa4UKk1YKJZAkzC5Sp7dzbeYIhmdcC6K7r9eXrdrnXgN6HR-1aBA3QC-cKXmQfVo-7XJT0HtqvNdNzGtrnbdww3NSHCrWirEFvZTQiTg~AxE4mn~xe7-Xmc-wRxHhTQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

Comments